8W1F
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer, Mg bound) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8W1E
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, SULFATE ION | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
7KEO
 
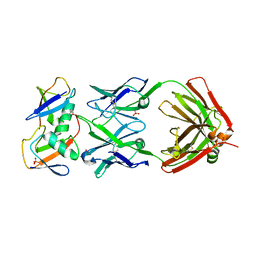 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
6JKV
 
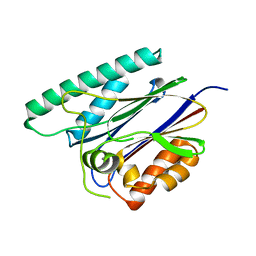 | | PppA, a key regulatory component of T6SS in Pseudomonas aeruginosa | | Descriptor: | MANGANESE (II) ION, PppA | | Authors: | Wang, T, Liu, L, Wu, Y, Li, D. | | Deposit date: | 2019-03-02 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PppA from Pseudomonas aeruginosa, a key regulatory component of type VI secretion systems.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6L8H
 
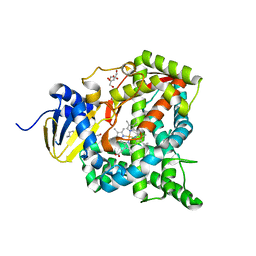 | | Crystal structure of CYP97C1 | | Descriptor: | Carotene epsilon-monooxygenase, chloroplastic, GLYCEROL, ... | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KZF
 
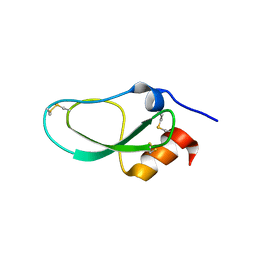 | | Racemic X-ray Structure of Calcicludine | | Descriptor: | D-calcicludine, Kunitz-type serine protease inhibitor homolog calcicludine | | Authors: | Qu, Q, Gao, S, Liu, L. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Synthesis of Disulfide Surrogate Peptides Incorporating Large-Span Surrogate Bridges Through a Native-Chemical-Ligation-Assisted Diaminodiacid Strategy
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L8J
 
 | | Crystal structure of CYP97A3 mutant S290D/W300L/S304V in complex with retinal | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic, ... | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L1G
 
 | |
4QDJ
 
 | |
4QDK
 
 | |
6MCT
 
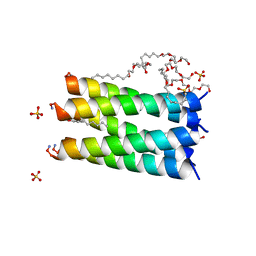 | |
4QS7
 
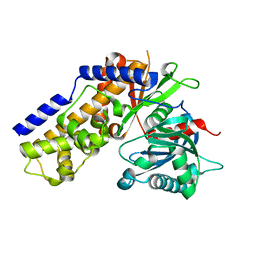 | |
6M09
 
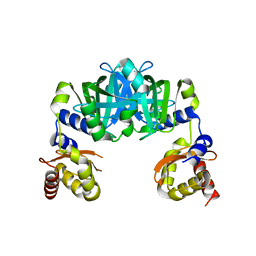 | |
4QS8
 
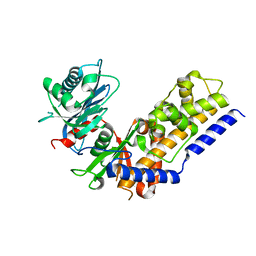 | |
8WB2
 
 | | Heme-bound Arabidopsis thaliana temperature-induced lipocalin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Temperature-induced lipocalin-1 | | Authors: | Dong, C, Liu, L. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and functional studies of a plant temperature-induced lipocalin.
Biochim Biophys Acta Gen Subj, 1868, 2023
|
|
4QS9
 
 | |
6MPW
 
 | |
8I6Y
 
 | | Crystal structure of Arabidopsis thaliana LOX1 | | Descriptor: | FE (III) ION, Linoleate 9S-lipoxygenase 1 | | Authors: | Liu, X, Liu, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | UV-B light signal mediates stomatal closure by activating the 9-lipoxygenase pathway
To Be Published
|
|
6NV1
 
 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, ... | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-02-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
4RIX
 
 | | Crystal structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-Q790R mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Littlefield, P, Liu, L, Jura, N. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Sci.Signal., 7, 2014
|
|
4RIY
 
 | | Crystal structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-E909G mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Littlefield, P, Liu, L, Jura, N. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Sci.Signal., 7, 2014
|
|
6M0A
 
 | | The heme-bound structure of the chloroplast protein At3g03890 | | Descriptor: | AT3G03890 protein, AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Liu, L. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Arabidopsis locus AT3G03890 encodes a dimeric beta-barrel protein implicated in heme degradation.
Biochem.J., 2020
|
|
6MQ2
 
 | |
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GM8
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-102 | | Descriptor: | MM-102, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
