5BSE
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, Pyrroline-5-carboxylate reductase | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
3BV6
 
 | | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold | | Descriptor: | FE (III) ION, Metal-dependent hydrolase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Yang, X, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold.
To be Published
|
|
5BSG
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NADP+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5CRF
 
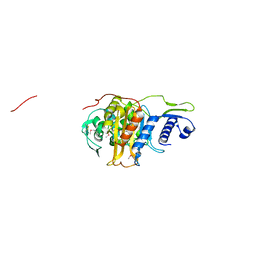 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
2BJO
 
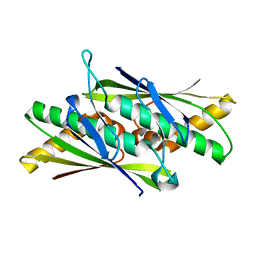 | | Crystal Structure of the Organic Hydroperoxide Resistance Protein OhrB of Bacillus subtilis | | Descriptor: | ORGANIC HYDROPEROXIDE RESISTANCE PROTEIN OHRB | | Authors: | Cooper, D.R, Surendranath, Y, Bielnicki, J, Devedjiev, Y, Joachimiak, A, Derewenda, Z.S. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Bacillus Subtilis Ohrb Hydroperoxide-Resistance Protein in a Fully Oxidized State.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6MDQ
 
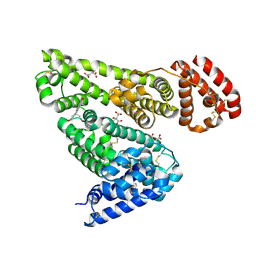 | | Crystal structure of equine serum albumin in complex with testosterone | | Descriptor: | CITRATE ANION, Serum albumin, TESTOSTERONE | | Authors: | Czub, M.P, Majorek, K.A, Shabalin, I.G, Handing, K.B, Venkataramany, B.S, Cymborowski, M.T, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-05 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Testosterone meets albumin - the molecular mechanism of sex hormone transport by serum albumins.
Chem Sci, 10, 2019
|
|
2AP1
 
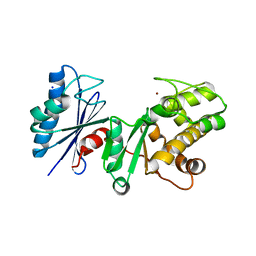 | | Crystal structure of the putative regulatory protein | | Descriptor: | SODIUM ION, ZINC ION, putative regulator protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative regulatory protein
To be Published
|
|
2AUA
 
 | | Structure of BC2332: A Protein of Unknown Function from Bacillus cereus | | Descriptor: | hypothetical protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-27 | | Release date: | 2005-10-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of BC2332: A hypothetical protein from Bacillus cereus
To be Published
|
|
2ETX
 
 | | Crystal Structure of MDC1 Tandem BRCT Domains | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Wasielewski, E, Kim, Y, Joachimiak, A, Thompson, J.R, Mer, G. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Basis for the Association of Microcephalin (MCPH1) Protein with the Cell Division Cycle Protein 27 (Cdc27) Subunit of the Anaphase-promoting Complex.
J.Biol.Chem., 287, 2012
|
|
3L9A
 
 | | Structure of the C-terminal domain from a Streptococcus mutans hypothetical | | Descriptor: | GLYCEROL, SODIUM ION, uncharacterized protein | | Authors: | Singer, A.U, Cuff, M.E, Xu, X, Cui, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-04 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the C-terminal domain from a Streptococcus mutans hypothetical
To be Published
|
|
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
3CNV
 
 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
1OEL
 
 | |
6C8Q
 
 | | Crystal structure of NAD synthetase (NadE) from Enterococcus faecalis in complex with NAD+ | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | To be published
To Be Published
|
|
3NRT
 
 | | The crystal structure of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative ryanodine receptor | | Authors: | Wu, R, Zhang, R, James, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | he crystal strucutre of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482
To be Published
|
|
3FUY
 
 | | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS1 | | Descriptor: | Putative integron gene cassette protein, SULFATE ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudrytska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-15 | | Release date: | 2009-02-24 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites
Plos One, 8, 2013
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3FY6
 
 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS3 | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-10 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
6CI6
 
 | | Crystal structure of equine serum albumin in complex with nabumetone | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Serum albumin, ... | | Authors: | Venkataramany, B.S, Czub, M.P, Shabalin, I.G, Handing, K.B, Steen, E.H, Cooper, D.R, Joachimiak, A, Satchell, K.J.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 2020
|
|
3GK6
 
 | | Crystal structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS2. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Integron cassette protein VCH_CASS2 | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Integron Gene Cassette-Associated Protein from Vibrio cholerae Identifies a Cationic Drug-Binding Module.
Plos One, 6, 2011
|
|
4KOR
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 7-aminocephalosporanic acid | | Descriptor: | 1,2-ETHANEDIOL, 7-aminocephalosporanic acid, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOV
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefuroxime | | Descriptor: | (6R,7R)-3-[(carbamoyloxy)methyl]-7-{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOU
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefixime | | Descriptor: | (6R,7R)-7-({(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-[(carboxymethoxy)imino]acetyl}amino)-3-ethenyl-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4L8A
 
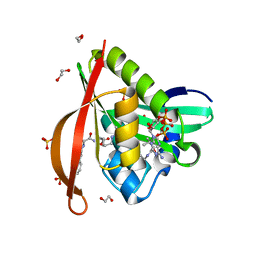 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in ternary complex with N-Phenylacetyl-Gly-AcLys and CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-(phenylacetyl)glycyl-N~6~-acetyl-L-lysine, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-06-16 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
4L89
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with covalently bound CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Maclean, E, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-06-16 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
