4R3B
 
 | |
3G7L
 
 | | Chromodomain of Chp1 in complex with Histone H3K9me3 peptide | | Descriptor: | ACETIC ACID, Chromo domain-containing protein 1, Histone H3.1/H3.2, ... | | Authors: | Schalch, T, Joshua-Tor, L. | | Deposit date: | 2009-02-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin
Mol.Cell, 34, 2009
|
|
4R8M
 
 | | Human SIRT2 crystal structure in complex with BHJH-TM1 | | Descriptor: | BHJH-TM1 peptide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Teng, Y.B, Hao, Q, Lin, H.N, Jing, H. | | Deposit date: | 2014-09-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Efficient Demyristoylase Activity of SIRT2 Revealed by Kinetic and Structural Studies
Sci Rep, 5, 2015
|
|
7UVR
 
 | | Crystal structure of human ClpP protease in complex with TR-65 | | Descriptor: | 3-{[(10R)-4-[(4-chlorophenyl)methyl]-5-oxo-1,2,4,5,8,9-hexahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-7(6H)-yl]methyl}benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UIB
 
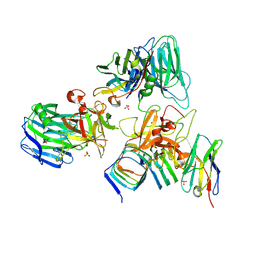 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2, VHH, and sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-beta-neuraminic acid, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
7UIA
 
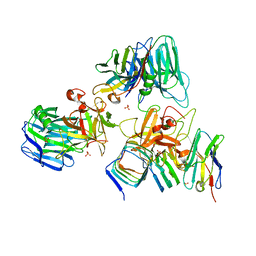 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2 and VHH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
7UIE
 
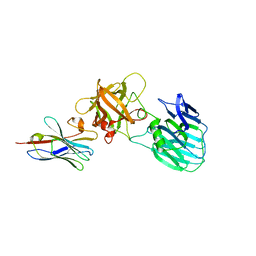 | | Crystal structure of HcE-JLE-G6 | | Descriptor: | Botulinum neurotoxin E heavy chain, JLE-G6 | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
3DNV
 
 | | MDT Protein | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3'), HTH-type transcriptional regulator hipB, Protein hipA, ... | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
7XJX
 
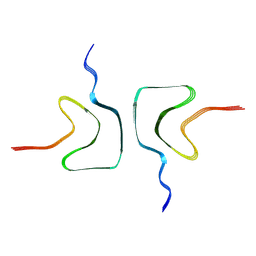 | | The cryo-EM structure of Fe3+ induced alpha-syn fibril. | | Descriptor: | Alpha-synuclein | | Authors: | Zhao, Q.Y, Tao, Y.Q, Zhao, K, Tao, Y.Q, Li, D. | | Deposit date: | 2022-04-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Insights of Fe3+ Induced alpha-synuclein Fibrillation in Parkinson' Disease
J.Mol.Biol., 435, 2023
|
|
7X27
 
 | | MERS-CoV spike complex | | Descriptor: | Spike glycoprotein | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | cryo-EM structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein
To Be Published
|
|
1PME
 
 | |
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3DJ4
 
 | | Crystal Structure of GlmU from Mycobacterium tuberculosis in complex with URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE. | | Descriptor: | Bifunctional protein glmU, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Verma, S.K, Prakash, B. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity.
J.Mol.Biol., 386, 2009
|
|
4RUD
 
 | | Crystal structure of a three finger toxin | | Descriptor: | Three-finger toxin 3b, ZINC ION | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2014-11-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fulditoxin, representing a new class of dimeric snake toxins, defines novel pharmacology at nicotinic ACh receptors.
Br.J.Pharmacol., 177, 2020
|
|
3VZE
 
 | |
4KS5
 
 | | Influenza neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
2MQ1
 
 | | Phosphotyrosine binding domain | | Descriptor: | E3 ubiquitin-protein ligase Hakai, ZINC ION | | Authors: | Mukherjee, M, Jing-Song, F, Sivaraman, J. | | Deposit date: | 2014-06-11 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dimeric switch of Hakai-truncated monomers during substrate recognition: insights from solution studies and NMR structure.
J.Biol.Chem., 289, 2014
|
|
4LRK
 
 | |
3VY6
 
 | |
3VZF
 
 | |
3VY7
 
 | |
3VZG
 
 | |
3WOG
 
 | | Crystal structure plant lectin in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-12-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
2HZC
 
 | |
2NYA
 
 | | Crystal structure of the periplasmic nitrate reductase (NAP) from Escherichia coli | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM(VI) ION, ... | | Authors: | Jepson, B.J.N, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli
J.Biol.Chem., 282, 2007
|
|
