8OUH
 
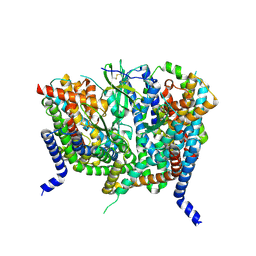 | | Complex of human ASCT2 with Syncytin-1 | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Syncytin-1 | | Authors: | Khare, S, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5DA0
 
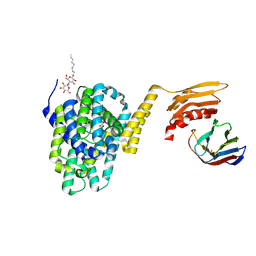 | | Structure of the the SLC26 transporter SLC26Dg in complex with a nanobody | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Nanobody, Sulphate transporter | | Authors: | Dutzler, R, Geertsma, E.R, Chang, Y, Shaik, F.R. | | Deposit date: | 2015-08-19 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6HLU
 
 | |
3GSP
 
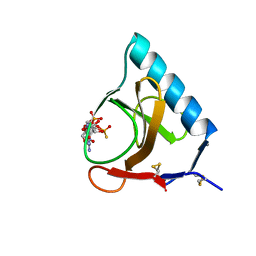 | | RIBONUCLEASE T1 COMPLEXED WITH 2',3'-CGPS + 3'-GMP, 4 DAYS | | Descriptor: | CALCIUM ION, GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, GUANOSINE-3'-MONOPHOSPHATE, ... | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-12-02 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
6HHD
 
 | | Mouse Prion Protein in complex with Nanobody 484 | | Descriptor: | CHLORIDE ION, GLYCEROL, Major prion protein, ... | | Authors: | Soror, S, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-28 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
6HER
 
 | | Mouse prion protein in complex with Nanobody 484 | | Descriptor: | GLYCEROL, Major prion protein, Nanobody 484, ... | | Authors: | Soror, S.H, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
3MKM
 
 | | Crystal structure of the E. coli pyrimidine nucleoside hydrolase YeiK (Apo-form) | | Descriptor: | CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
4AQ1
 
 | |
6H02
 
 | |
4CGZ
 
 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with DNA | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*CP*GP*AP*GP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*CP*TP*CP*GP*AP*CP*GP*CP*TP*CP*DT *CP*CP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Newman, J.A, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
9H76
 
 | | Bacterial LAT transporter BASC in complex with L-Ala and NB53 | | Descriptor: | ALANINE, Nanobody NB53, Putative amino acid/polyamine transport protein | | Authors: | Fort, J, Palacin, M. | | Deposit date: | 2024-10-26 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The conserved lysine residue in transmembrane helix 5 is pivotal for the cytoplasmic gating of the L-amino acid transporters.
Pnas Nexus, 4, 2025
|
|
4DK3
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4DKA
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, SODIUM ION, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
5IOF
 
 | |
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
4M3K
 
 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
6QNW
 
 | |
6QWL
 
 | | Influenza B virus (B/Panama/45) polymerase Hetermotrimer in complex with 3'5' cRNA promoter | | Descriptor: | 3' cRNA, 5' cRNA, Polymerase acidic protein, ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QPF
 
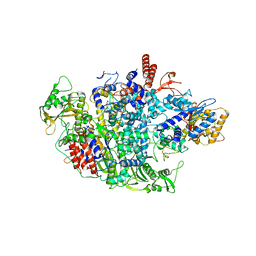 | | Influenza A virus Polymerase Heterotrimer A/duck/Fujian/01/2002(H5N1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.634 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
4M3J
 
 | | Structure of a single-domain camelid antibody fragment cAb-H7S specific of the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Camelid heavy-chain antibody variable fragment cAb-H7S, SULFATE ION | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
6QX3
 
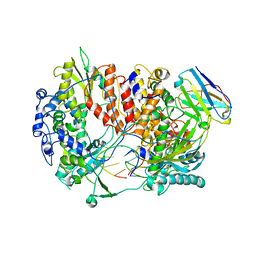 | | Influenza A virus (A/NT/60/1968) polymerase Hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
4N1H
 
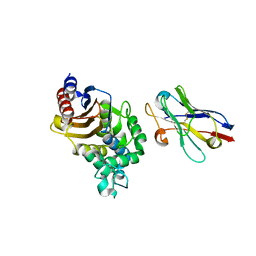 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
6RR7
 
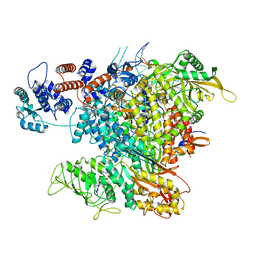 | | Influenza A virus (A/NT/60/1968) polymerase Heterotrimer bound to 3'5' vRNA promoter and capped RNA primer | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(M7G))-R(P*AP*AP*UP*CP*U)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QPG
 
 | | Influenza A virus Polymerase Heterotrimer A/nt/60/1968(H3N2) in complex with Nanobody NB8205 | | Descriptor: | Nanobody NB8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6QX8
 
 | | Influenza A virus (A/NT/60/1968) polymerase dimer of heterotrimer in complex with 5' cRNA promoter | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*A)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
