3JS4
 
 | |
3P85
 
 | |
3K2H
 
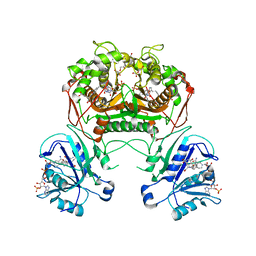 | | Co-crystal structure of dihydrofolate reductase/thymidylate synthase from Babesia bovis with dUMP, Pemetrexed and NADP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor-bound complexes of dihydrofolate reductase-thymidylate synthase from Babesia bovis.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3P4I
 
 | |
3OC9
 
 | |
3O38
 
 | |
3JS5
 
 | |
3JVI
 
 | |
3OXK
 
 | |
3P08
 
 | |
3PZY
 
 | |
3JS9
 
 | |
3PM6
 
 | |
3QLJ
 
 | |
3QXI
 
 | |
3QDF
 
 | |
3QXZ
 
 | |
3MEN
 
 | |
3PY5
 
 | |
3MOY
 
 | |
3QKA
 
 | |
3QBP
 
 | |
3QMJ
 
 | |
3QH8
 
 | |
3QD5
 
 | |
