8W8F
 
 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8W8E
 
 | | human co-transcriptional RNA capping enzyme RNGTT | | Descriptor: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8W56
 
 | | Cryo-EM structure of DSR2-DSAD1 state 1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-08-25 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8W7J
 
 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC with chain A bounded to substrate PneA and GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, PneA LP, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J, Li, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8IN8
 
 | | Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*A)-3'), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8WKN
 
 | | Cryo-EM structure of DSR2-DSAD1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
4I79
 
 | | Crystal structure of human NUP43 | | Descriptor: | Nucleoporin Nup43, UNKNOWN ATOM OR ION, floating chain, ... | | Authors: | Xu, C, Tempel, W, Li, Z, He, H, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-11-30 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of human nuclear pore complex component NUP43.
Febs Lett., 589, 2015
|
|
5A9Y
 
 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9X
 
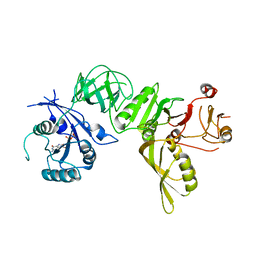 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6V05
 
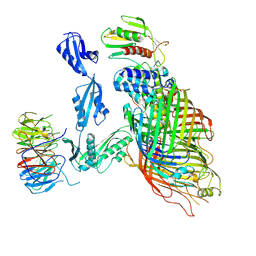 | | Cryo-EM structure of a substrate-engaged Bam complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
5A9W
 
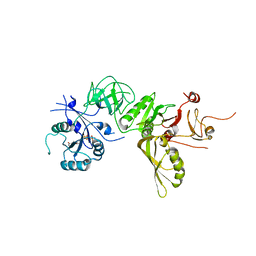 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
5A6F
 
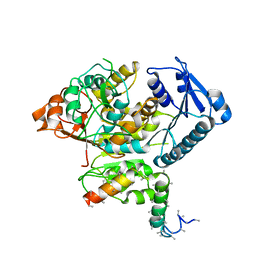 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A22
 
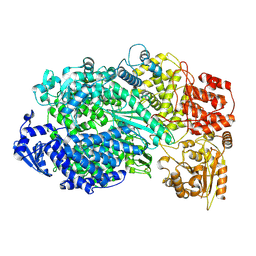 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | Descriptor: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | Authors: | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
8WFN
 
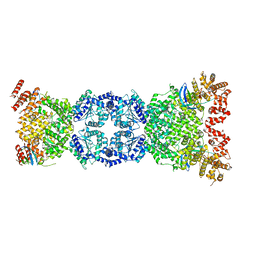 | | Cryo-EM structure of DSR2-TTP | | Descriptor: | SIR2-like domain-containing protein, tail tube protein(TTP) | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Cryo-EM structure of DSR2-TTP
To Be Published
|
|
8WCK
 
 | | FCP tetramer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, Chlorophyll a/b-binding protein, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Shen, J.-R, Liu, C, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 5, 2024
|
|
8WCL
 
 | | FCP pentamer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Liu, C, Shen, J.-R, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 5, 2024
|
|
8W9X
 
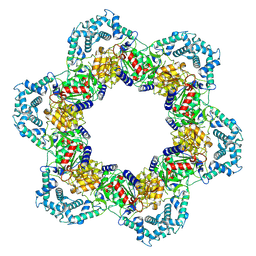 | |
8W9L
 
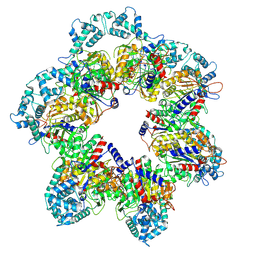 | |
5A6E
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, ... | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
6W5C
 
 | | Cryo-EM structure of Cas12i(E894A)-crRNA-dsDNA complex | | Descriptor: | Cas12i, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5A9V
 
 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
