7TJH
 
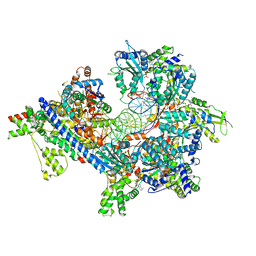 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJK
 
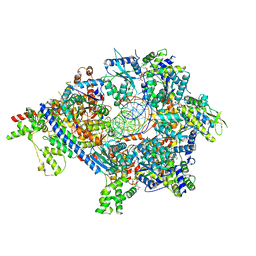 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJJ
 
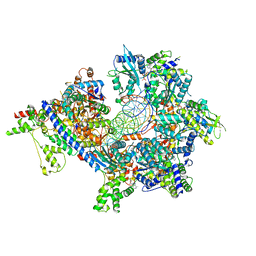 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with docked Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJF
 
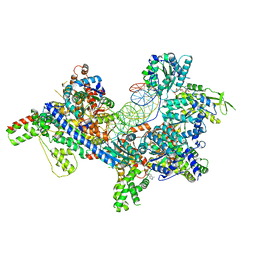 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA, 84 bp ARS1, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-18 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TVI
 
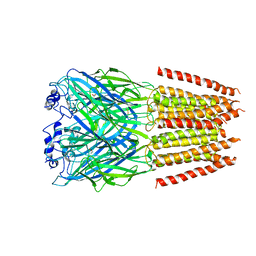 | | Alpha1/BetaB Heteromeric Glycine Receptor in Glycine-Bound State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor beta subunit 2, ... | | Authors: | Gibbs, E, Chakrapani, S, Kumar, A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-03-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and allosteric modulation in a heteromeric glycine receptor
Nat Commun, 14, 2023
|
|
7TU9
 
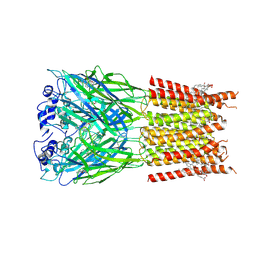 | | Alpha1/BetaB Heteromeric Glycine Receptor in Strychnine-Bound State | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor beta subunit 2, ... | | Authors: | Gibbs, E, Kumar, A, Chakrapani, S. | | Deposit date: | 2022-02-02 | | Release date: | 2023-03-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions and allosteric modulation in a heteromeric glycine receptor
Nat Commun, 14, 2023
|
|
7YH4
 
 | |
1VZK
 
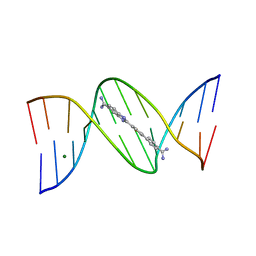 | | A Thiophene Based Diamidine Forms a "Super" AT Binding Minor Groove Agent | | Descriptor: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-THIENYL)-1H-BENZIMIDAZOLE-6- CARBOXIMIDAMIDE DIHYDROCHLORIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP *CP*G)-3', MAGNESIUM ION | | Authors: | Mallena, S, Lee, M.P.H, Bailly, C, Neidle, S, Kumar, A, Boykin, D.W, Wilson, W.D. | | Deposit date: | 2004-05-20 | | Release date: | 2004-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thiophene-Based Diamidine Forms a "Super" at Binding Minor Groove Agent
J.Am.Chem.Soc., 142, 2004
|
|
4QAJ
 
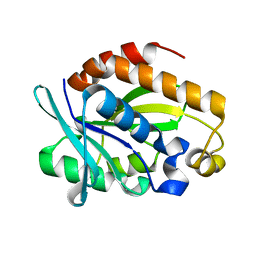 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4JC4
 
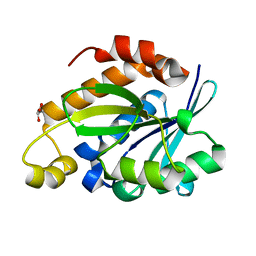 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2013-02-21 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
5GIQ
 
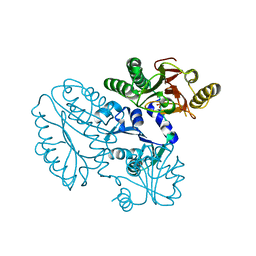 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | Authors: | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
5GIV
 
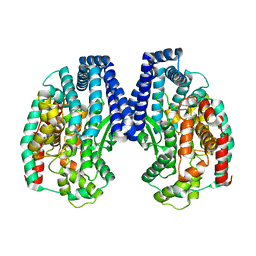 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | Authors: | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
6M1C
 
 | |
7CLE
 
 | | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS | | Descriptor: | Acid phosphatase, MAGNESIUM ION | | Authors: | Gaur, N.K, Kumar, A, Sunder, S, Mukhopadhyaya, R, Makde, R.D. | | Deposit date: | 2020-07-20 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS
To Be Published
|
|
7CZC
 
 | | Crystal structure of apo-FabG from Vibrio harveyi | | Descriptor: | 3-oxoacyl-ACP reductase FabG, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, B.K, Kumar, A, Paul, B, Biswas, R, Das, A.K. | | Deposit date: | 2020-09-08 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of apo-FabG from Vibrio harveyi
To Be Published
|
|
7CAY
 
 | | Crystal Structure of Lon N-terminal domain protein from Xanthomonas campestris | | Descriptor: | ATP-dependent protease | | Authors: | Singh, R, Sharma, B, Deshmukh, S, Kumar, A, Makde, R.D. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5CAD
 
 | | Crystal structure of the vicilin from Solanum melongena revealed existence of different anionic ligands in structurally similar pockets | | Descriptor: | ACETATE ION, MAGNESIUM ION, PYROGLUTAMIC ACID, ... | | Authors: | Jain, A, Kumar, A, Salunke, D.M. | | Deposit date: | 2015-06-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the vicilin from Solanum melongena reveals existence of different anionic ligands in structurally similar pockets
Sci Rep, 6, 2016
|
|
1FMQ
 
 | | Cyclo-butyl-bis-furamidine complexed with dCGCGAATTCGCG | | Descriptor: | 2,5-BIS{[4-(N-CYCLOBUTYLDIAMINOMETHYL)PHENYL]}FURAN, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Simpson, I.J, Lee, M, Kumar, A, Boykin, D.W, Neidle, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA minor groove interactions and the biological activity of 2,5-bis-[4-(N-alkylamidino)phenyl] furans
Bioorg.Med.Chem.Lett., 10, 2000
|
|
1FMS
 
 | | Structure of complex between cyclohexyl-bis-furamidine and d(CGCGAATTCGCG) | | Descriptor: | 2,5-BIS{[4-(N-CYCLOHEXYLDIAMINOMETHYL)PHENYL]}FURAN, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Simpson, I.J, Lee, M, Kumar, A, Boykin, D.W, Neidle, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA minor groove interactions and the biological activity of 2,5-bis-[4-(N-alkylamidino)phenyl] furans
Bioorg.Med.Chem.Lett., 10, 2000
|
|
6JBP
 
 | | Structure of MP-4 from Mucuna pruriens at 2.22 Angstroms | | Descriptor: | Kunitz-type trypsin inhibitor-like 2 protein | | Authors: | Jain, A, Shikhi, M, Kumar, A, Kumar, A, Nair, D.T, Salunke, D.M. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | The structure of MP-4 from Mucuna pruriens at 2.22 angstrom resolution.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2LGJ
 
 | | Solution structure of MsPTH | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of Peptidyl t-RNA hydrolase from Mycobacterium smegmatis
To be Published
|
|
5ULM
 
 | |
5CE6
 
 | | N-terminal domain of FACT complex subunit SPT16 from Cicer arietinum (chickpea) | | Descriptor: | ACETATE ION, FACT-Spt16, POTASSIUM ION, ... | | Authors: | Are, V.N, Ghosh, B, Kumar, A, Makde, R. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and dynamics of Spt16N-domain of FACT complex from Cicer arietinum.
Int.J.Biol.Macromol., 88, 2016
|
|
4JHW
 
 | | Crystal Structure of Respiratory Syncytial Virus Fusion Glycoprotein Stabilized in the Prefusion Conformation by Human Antibody D25 | | Descriptor: | D25 antigen-binding fragment heavy chain, D25 light chain, Fusion glycoprotein F0 | | Authors: | Mclellan, J.S, Chen, M, Leung, S, Graepel, K.W, Du, X, Yang, Y, Zhou, T, Baxa, U, Yasuda, E, Beaumont, T, Kumar, A, Modjarrad, K, Zheng, Z, Zhao, M, Xia, N, Kwong, P.D, Graham, B.S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Science, 340, 2013
|
|
4P8E
 
 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
