4C9R
 
 | |
4C99
 
 | |
4C8F
 
 | | mouse ZNRF3 ectodomain crystal form IV | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C9A
 
 | |
2CL8
 
 | | Dectin-1 in complex with beta-glucan | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECTIN-1, ... | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2006-04-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BPD
 
 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1 | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BPH
 
 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1, MAGNESIUM ION | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BVE
 
 | | Structure of the N-terminal of Sialoadhesin in complex with 2-Phenyl- Prop5Ac | | Descriptor: | SIALOADHESIN, benzyl 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosidonic acid | | Authors: | Zaccai, N.R, May, A.P, Robinson, R.C, Burtnick, L.D, Crocker, P, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2005-06-27 | | Release date: | 2006-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and in Silico Analysis of the Sialoside-Binding Characteristics of the Siglec Sialoadhesin.
J.Mol.Biol., 365, 2007
|
|
2BZX
 
 | |
7B3G
 
 | | Notum complex with ARUK3003902 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-((2-chlorophenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
2CII
 
 | | The crystal structure of H-2Db complexed with a partial peptide epitope suggests an MHC Class I assembly-intermediate | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN D-B ALPHA CHAIN, ... | | Authors: | Glithero, A, Tormo, J, Doering, K, Kojima, M, Jones, E.Y, Elliott, T. | | Deposit date: | 2006-03-21 | | Release date: | 2006-03-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of H-2D(b) complexed with a partial peptide epitope suggests a major histocompatibility complex class I assembly intermediate.
J. Biol. Chem., 281, 2006
|
|
2CDG
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2BPE
 
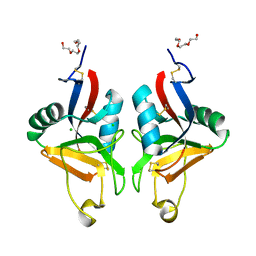 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECTIN-1, ... | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
4B7I
 
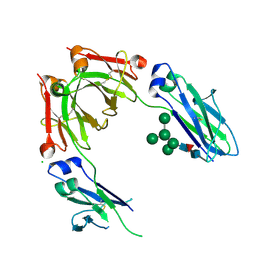 | | Crystal Structure of Human IgG Fc Bearing Hybrid-type Glycans | | Descriptor: | CHLORIDE ION, IG GAMMA-1 CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bowden, T.A, Baruah, K, Coles, C.H, Harvey, D.J, Song, B.D, Stuart, D.I, Aricescu, A.R, Scanlan, C.N, Jones, E.Y, Crispin, M. | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Chemical and Structural Analysis of an Antibody Folding Intermediate Trapped During Glycan Biosynthesis.
J.Am.Chem.Soc., 134, 2012
|
|
4C84
 
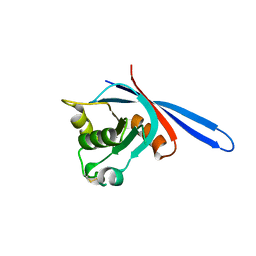 | | zebrafish ZNRF3 ectodomain crystal form I | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
2BRY
 
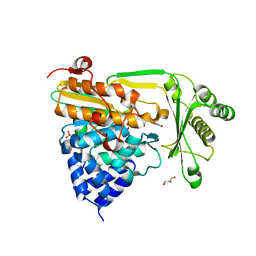 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-05-13 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C4C
 
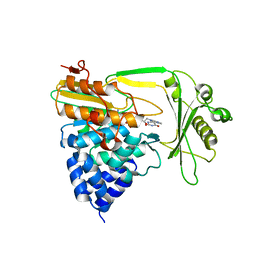 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-10-18 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C9A
 
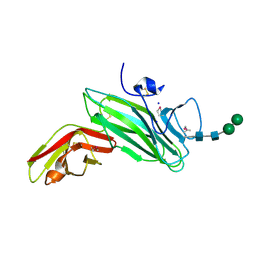 | | Crystal structure of the MAM-Ig module of receptor protein tyrosine phosphatase mu | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE MU, ... | | Authors: | Aricescu, A.R, Hon, W.C, Siebold, C, Lu, W, Van Der Merwe, P.A, Jones, E.Y. | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Analysis of Receptor Protein Tyrosine Phosphatase Mu-Mediated Cell Adhesion.
Embo J., 25, 2006
|
|
2BZY
 
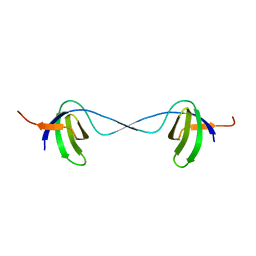 | |
7B2Z
 
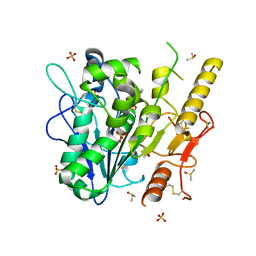 | | Notum complex with ARUK3003907 | | Descriptor: | DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, SULFATE ION, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B37
 
 | | Notum complex with ARUK3003718 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2,3-dihydroindol-1-yl)-4-oxidanylidene-butanoic acid, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B2Y
 
 | | Notum complex with ARUK3003910 | | Descriptor: | 1,2-ETHANEDIOL, 2,2-bis(fluoranyl)ethyl 4-(2,3-dihydroindol-1-yl)-4-oxidanylidene-butanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B2V
 
 | | Notum complex with ARUK3003906 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
7B3F
 
 | | Notum S232A in complex with ARUK3003718 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2,3-dihydroindol-1-yl)-4-oxidanylidene-butanoic acid, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-30 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into Notum Covalent Inhibition.
J.Med.Chem., 64, 2021
|
|
4BKA
 
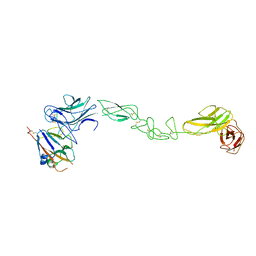 | | crystal structure of the human EphA4 ectodomain in complex with human ephrin A5 | | Descriptor: | EPHRIN TYPE-A RECEPTOR 4, EPHRIN-A5 | | Authors: | Seiradake, E, Schaupp, A, del Toro Ruiz, D, Kaufmann, R, Mitakidis, N, Harlos, K, Aricescu, A.R, Klein, R, Jones, E.Y. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.3 Å) | | Cite: | Structurally Encoded Intraclass Differences in Epha Clusters Drive Distinct Cell Responses
Nat.Struct.Mol.Biol., 20, 2013
|
|
