3G3D
 
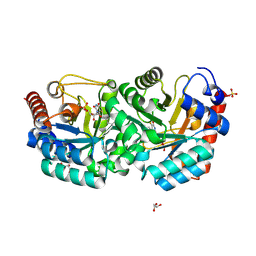 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-azido-UMP | | Descriptor: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, Y, Tang, H.L, Bello, A, Poduch, E, Kotra, L, Pai, E. | | Deposit date: | 2009-02-02 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
4QQG
 
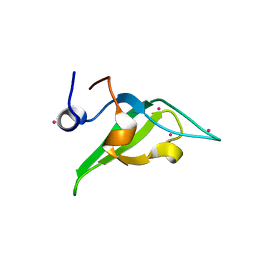 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
5H06
 
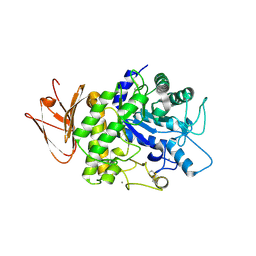 | | Crystal structure of AmyP in complex with maltose | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
3S9Y
 
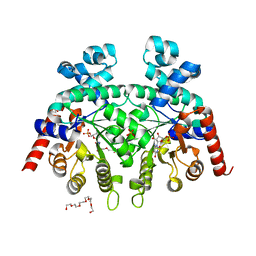 | | Crystal Structure of P. falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP in space group P21, produced from 5-fluoro-6-azido-UMP | | Descriptor: | 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
6MLC
 
 | | PHD6 domain of MLL3 in complex with histone H4 | | Descriptor: | GLYCEROL, Histone H4, Histone-lysine N-methyltransferase 2C, ... | | Authors: | Dong, A, Liu, Y, Qin, S, Lei, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4.
Nat Commun, 10, 2019
|
|
4PLL
 
 | |
4RGY
 
 | |
2FYZ
 
 | |
5IN1
 
 | | Crystal Structure of the MRG701 chromodomain | | Descriptor: | 1,2-ETHANEDIOL, MRG701, SULFATE ION | | Authors: | Huang, Y, Liu, Y. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on MRG701 chromodomain reveal a novel dimerization interface of MRG proteins in green plants
Protein Cell, 7, 2016
|
|
4PLI
 
 | |
5H05
 
 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
5EPL
 
 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
2QAF
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase covalently modified by 6-iodo-UMP | | Descriptor: | Orotidine 5' monophosphate decarboxylase, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | Deposit date: | 2007-06-15 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-activity relationships of C6-uridine derivatives targeting plasmodia orotidine monophosphate decarboxylase
J.Med.Chem., 51, 2008
|
|
2Q8Z
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase complexed with 6-amino-UMP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 6-AMINOURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of C6-Uridine Derivatives Targeting Plasmodia Orotidine Monophosphate Decarboxylase.
J.Med.Chem., 51, 2008
|
|
5W3L
 
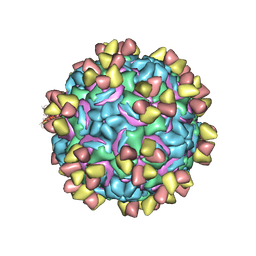 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3E
 
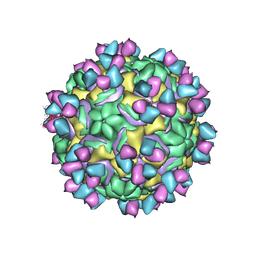 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5EPK
 
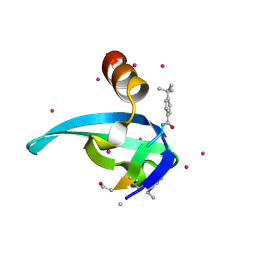 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EG8
 
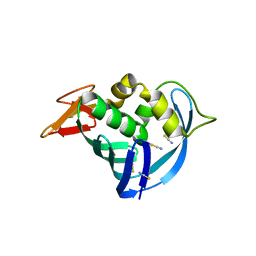 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2, THIOCYANATE ION | | Authors: | Liu, Y, Zheng, X, Severin, C, Rocha de Moura, T, Li, K, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EQ0
 
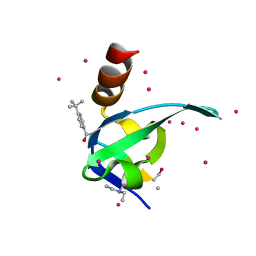 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
8K7O
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8I
 
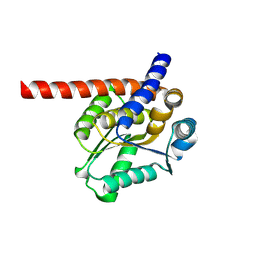 | | De novo design protein -N14 | | Descriptor: | De novo design protein -N14 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8F
 
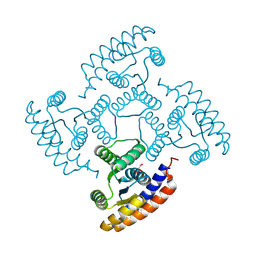 | | De novo design protein -N7 | | Descriptor: | De novo design protein N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7Z
 
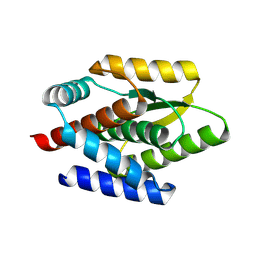 | | De novo design protein -N1 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7M
 
 | | De novo design protein -T01 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K83
 
 | | De novo design protein -N2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, De novo design protein, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
