9GPQ
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12h/p | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-17-methyl-11,18-dioxa-1,17lambda5-diphospha-19lambda5-stannapentacyclo[17.3.1.04,9.04,13.015,20]tricosane-2,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPX
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12f/l | | Descriptor: | 3-cyclohexyl-17-hydroxy-16,17-dimethoxy-12-methyl-15,18-dioxa-5-aza-1lambda5,17lambda5-diphosphapentacyclo[17.3.1.01,10.05,10.011,19]tricosa-10,19(23)-diene-4,11-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7PY4
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dieudonne, T, Abad-Herrera, S, Juknaviciute Laursen, M, Lejeune, M, Stock, C, Slimani, K, Jaxel, C, Lyons, J.A, Montigny, C, Gunther Pomorski, T, Nissen, P, Lenoir, G. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Autoinhibition and regulation by phosphoinositides of ATP8B1, a human lipid flippase associated with intrahepatic cholestatic disorders.
Elife, 11, 2022
|
|
9GPW
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12f/j | | Descriptor: | 2-cyclohexyl-18,19-dimethoxy-13-methyl-11,17-dioxa-4-aza-1lambda5,18lambda5-diphospha-1,15-distannatetracyclo[13.4.0.04,9.016,20]nonadecane-3,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPL
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 11c/i | | Descriptor: | 2-cyclohexyl-1,19-dimethoxy-11,18-dioxa-1lambda5,19lambda5-diphospha-2-stannatetracyclo[15.3.0.01,13.04,9]icosane-3,12-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPK
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/j | | Descriptor: | 2-cyclohexyl-1,19-dimethoxy-11,18-dioxa-1lambda5,19lambda5-diphospha-2-stannatetracyclo[18.2.1.01,6.06,10]tricosane-3,12-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Walz, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPR
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12h/m | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-17-methyl-11,15-dioxa-19lambda4-thia-5-aza-18-stannapentacyclo[15.7.0.01,10.010,10.018,23]tetracosa-13,20-diene-4,12-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPO
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/n | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-17-methyl-11,15-dioxa-1-aza-1lambda5-polona-1,18-distannapentacyclo[15.7.0.01,10.010,10.011,16]eicosane-2,12-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPT
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/k | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-19-methyl-11,18-dioxa-1,19lambda5-dithia-5-aza-19lambda5-stannapentacyclo[18.3.1.04,9.013,22.015,21]tetracosane-2,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPY
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12g/j | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-18,19-dimethoxy-13-methyl-11,17-dioxa-4-aza-1lambda5,18lambda5-diphospha-1,15-distannatetracyclo[13.4.0.04,9.016,20]nonadecane-3,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPZ
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12f/o | | Descriptor: | 3-cyclohexyl-17,18-dimethoxy-13-methyl-17-methylol-15,19-dioxa-1,17lambda5-diphospha-1,11lambda5-distannapentacyclo[15.3.1.04,9.011,13.013,19]heneicosane-2,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | Fab S24-1379, heavy chain, light chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7SUE
 
 | | Crystal Structure of Human Fab S24-188 in the complex with the N-teminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | Nucleoprotein, S24-188 Fab Heavy chain, S24-188 Fab Light chain | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
8BT9
 
 | |
6Y94
 
 | | Ca2+-bound Calmodulin mutant N53I | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Holt, C, Nielsen, L.H, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6Y4P
 
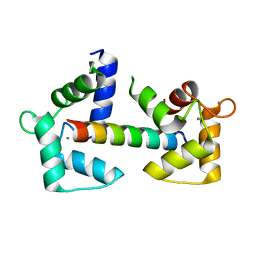 | | Calmodulin N53I variant bound to cardiac ryanodine receptor (RyR2) calmodulin binding domain | | Descriptor: | CALCIUM ION, Calmodulin-1, Ryanodine receptor 2 | | Authors: | Lau, K, Nielsen, L.H, Holt, C, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Van Petegem, F, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13325572 Å) | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
8K2S
 
 | |
1AFV
 
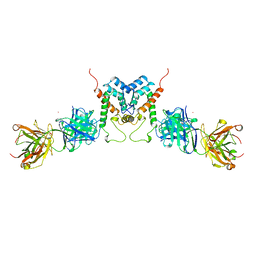 | | HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 | | Descriptor: | ANTIBODY FAB25.3 FRAGMENT (HEAVY CHAIN), ANTIBODY FAB25.3 FRAGMENT (LIGHT CHAIN), HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 CAPSID PROTEIN, ... | | Authors: | Momany, C, Kovari, L.C, Prongay, A.J, Keller, W, Gitti, R.K, Lee, B.M, Gorbalenya, A.E, Tong, L, Mcclure, J, Ehrlich, L.S, Summers, M.F, Carter, C, Rossmann, M.G. | | Deposit date: | 1997-03-14 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of dimeric HIV-1 capsid protein.
Nat.Struct.Biol., 3, 1996
|
|
1CUY
 
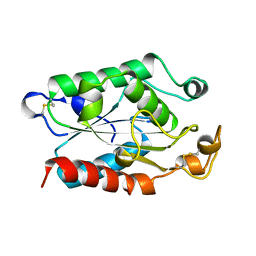 | | CUTINASE, L189F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CEX
 
 | | STRUCTURE OF CUTINASE | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Czjzek, M, Lamzin, V, Nicolas, A, Cambillau, C. | | Deposit date: | 1997-02-18 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: stereochemical analysis.
J.Mol.Biol., 268, 1997
|
|
1NAM
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
8K2R
 
 | |
6G4Q
 
 | | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2 | | Descriptor: | 1,2-ETHANEDIOL, Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial, ... | | Authors: | Bailey, H.J, Shrestha, L, Rembeza, E, Sorrell, F.J, Newman, J, Strain-Damerell, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2
To Be Published
|
|
8K2T
 
 | |
