8B14
 
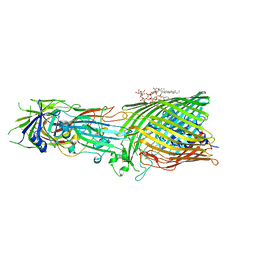 | | T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, FhuA iron-ferrichrome transporter, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate, ... | | Authors: | Degroux, S, Effantin, G, Linares, R, Schoehn, G, Breyton, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger.
J.Virol., 97, 2023
|
|
5YQZ
 
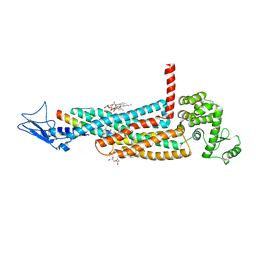 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
8B9R
 
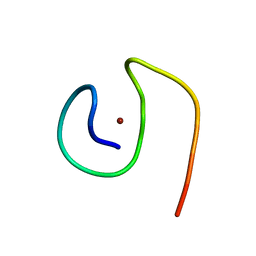 | | Molecular structure of Cu(II)-bound amyloid-beta monomer implicated in inhibition of peptide self-assembly in Alzheimer's disease | | Descriptor: | Amyloid-beta A4 protein, COPPER (II) ION | | Authors: | Abelein, A, Ciofi-Baffoni, S, Morman, C, Kumar, R, Giachetti, A, Piccioli, M, Biverstal, H. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Structure of Cu(II)-Bound Amyloid-beta Monomer Implicated in Inhibition of Peptide Self-Assembly in Alzheimer's Disease.
Jacs Au, 2, 2022
|
|
8AX2
 
 | |
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
8AWZ
 
 | |
8B8F
 
 | | Atomic structure of the beta-trefoil domain of the Laccaria bicolor lectin LBL in complex with lactose | | Descriptor: | N-terminal beta-trefoil domain of the lectin LBL from Laccaria bicolor, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
6XUB
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with monovinyl monopropionyl deuteroheme | | Descriptor: | Chlorite dismutase, harderoheme (III) | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
8B97
 
 | | N-terminal beta-trefoil lectin domain of the Laccaria bicolor lectin in complex with N-acetyl-lactosamine | | Descriptor: | Beta-trefoil domain of the LBL lectin, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
8G2O
 
 | | Fab structure - Anti-ApoE-7C11 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, anti-ApoE-7C11 heavy chain, anti-ApoE-7C11 light chain | | Authors: | Marino, C, Arboleda-Velasquez, J.F. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | APOE Christchurch-mimetic therapeutic antibody reduces APOE-mediated toxicity and tau phosphorylation.
Alzheimers Dement, 20, 2024
|
|
5L76
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in apo form) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Pena, I.A, Rembeza, E, Strain-Damerell, C, Chalk, R, Borkowska, O, Goubin, S, Velupillai, S, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Arruda, P, Yue, W.W. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in apo form)
To Be Published
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5A3W
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with Pyridine-2, 6-dicarboxylic Acid (PDCA) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
8GCM
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (5S)-5-[(3R)-4,4-difluoro-3-hydroxy-4-phenylbutyl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GCP
 
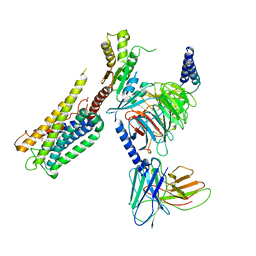 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KW2
 
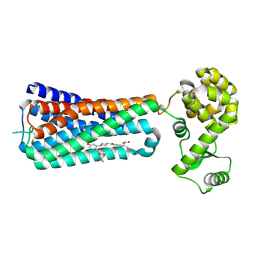 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
8GD9
 
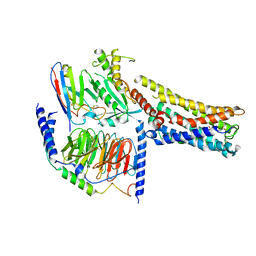 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | 4-({2-[(1R,2R,3R,5S)-3,5-dihydroxy-2-{(3S)-3-hydroxy-4-[3-(methoxymethyl)phenyl]butyl}cyclopentyl]ethyl}sulfanyl)butanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Yang, D, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4MMO
 
 | | The crystal structure of a M20 family metallo-carboxypeptidase Sso-CP2 from Sulfolobus solfataricus | | Descriptor: | GLYCEROL, SULFATE ION, Sso-CP2 metallo-carboxypetidase, ... | | Authors: | Dupuy, J, Dutoit, R, Durisotti, V, Demarez, M, Borel, F, Van Elder, D, Legrain, C, Bauvois, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3363 Å) | | Cite: | Biochemical characterization of a novel thermostable dinuclear carboxypeptidase from the thermoacidophilic archaeum Sulfolobus solfataricus.
To be Published
|
|
6ZXI
 
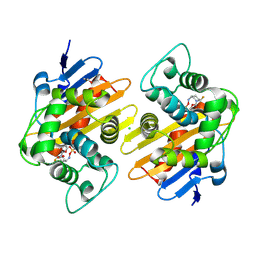 | | Crystal Structure of the OXA-48 Carbapenem-Hydrolyzing Class D beta-Lactamase in Complex with the DBO inhibitor ANT3310 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CARBON DIOXIDE, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ANT3310 , a Novel Broad-Spectrum Serine beta-Lactamase Inhibitor of the Diazabicyclooctane Class, Which Strongly Potentiates Meropenem Activity against Carbapenem-Resistant Enterobacterales and Acinetobacter baumannii.
J.Med.Chem., 63, 2020
|
|
9GPN
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 11b/j | | Descriptor: | 3-cyclohexyl-1,19-dimethoxy-11,16-dioxa-19lambda4-polona-1,13-diaza-2lambda5-polonapentacyclo[16.4.0.04,9.013,19.020,25]docosane-2,12-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPP
 
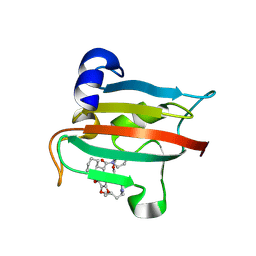 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/o | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-19-methyl-11,20-dioxa-5-aza-1lambda5-polona-1,19lambda5-dithia-18-azanida-1lambda5-stannapentacyclo[19.2.1.05,10.013,18.018,22]tetracosane-4,12-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPM
 
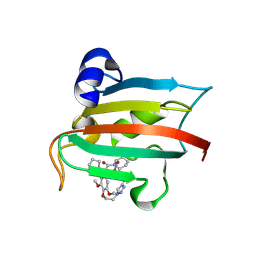 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/i | | Descriptor: | 3-cyclohexyl-17,18-dimethoxy-11,15-dioxa-1,18lambda4-dithia-5-aza-17lambda4-polonapentacyclo[15.4.0.04,9.013,18.018,23]henicosane-2,12-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPS
 
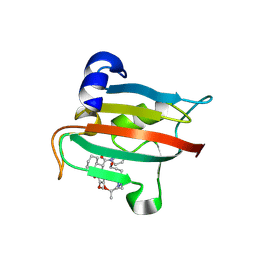 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/p | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-19-methyl-11,20-dioxa-5-aza-1lambda5-polona-1,19lambda5-dithia-18-azanida-1lambda5-stannapentacyclo[19.2.1.05,10.013,18.018,22]tetracosane-4,12-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPU
 
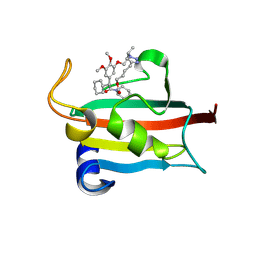 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12h/k | | Descriptor: | 3-cyclohexyl-23,24-dimethoxy-17-methyl-11,22-dioxa-5-aza-23lambda5,24lambda5-diphospha-2,23lambda5-diphosphapentacyclo[21.2.1.01,18.05,10.018,23]hexacosa-18,24-diene-4,12-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GPV
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 12c/l | | Descriptor: | 3-cyclohexyl-19,20-dimethoxy-19-methyl-11,18-dioxa-1,19lambda5-dithia-5-aza-19lambda5-stannapentacyclo[18.3.1.04,9.013,22.015,21]tetracosane-2,10-quinone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Walz, C, Hausch, F. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Plasticity and Binding Affinity Enhancement Controlled by Linker Derivatization in Macrocycles.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
