5KWI
 
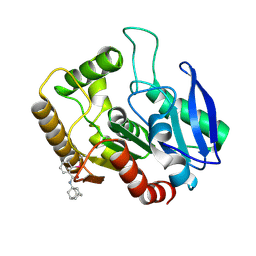 | | M.tb Ag85C modified at C209 by adamantyl-ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, ~{N}-(1-adamantyl)-2-selanyl-benzamide | | Authors: | Goins, C.M, Ronning, D.R. | | Deposit date: | 2016-07-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Exploring Covalent Allosteric Inhibition of Antigen 85C from Mycobacterium tuberculosis by Ebselen Derivatives.
ACS Infect Dis, 3, 2017
|
|
7R9C
 
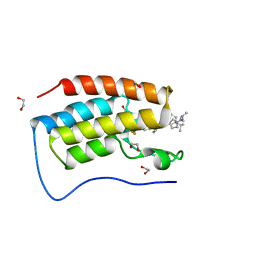 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
4ZW5
 
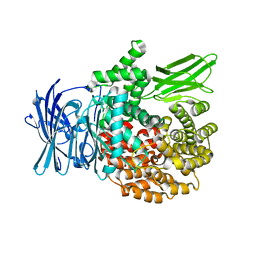 | |
4ZX5
 
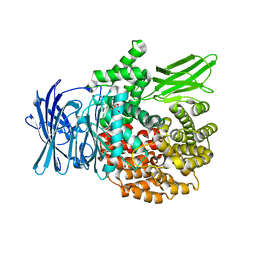 | |
4ZW6
 
 | |
2MD0
 
 | | Solution structure of ShK-like immunomodulatory peptide from Ancylostoma caninum (hookworm) | | Descriptor: | AcK1 | | Authors: | Chhabra, S, Swarbrick, J.D, Mohanty, B, Chang, S.C, Chandy, G.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
4JOA
 
 | | Crystal Structure of Human Anaplastic Lymphoma Kinase in complex with 7-azaindole based inhibitor | | Descriptor: | 3-[1-(2,5-difluorobenzyl)-1H-pyrazol-4-yl]-5-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, ALK tyrosine kinase receptor | | Authors: | Hosahalli, S, Krishnamurthy, N.R, Lakshminarasimhan, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-azaindole based anaplastic lymphoma kinase (ALK) inhibitors: wild type and mutant (L1196M) active compounds with unique binding mode
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8AXR
 
 | | Crystal structure of the C-terminal domain of human CFAP410 | | Descriptor: | CHLORIDE ION, Cilia- and flagella-associated protein 410 | | Authors: | Dong, G, Stadler, A. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminus of CFAP410 forms a tetrameric helical bundle that is essential for its localization to the basal body.
Open Biology, 14, 2024
|
|
8AXO
 
 | |
5UIX
 
 | |
6XCU
 
 | |
5WKH
 
 | | D30 TCR in complex with HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS3 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WKF
 
 | | D30 TCR in complex with HLA-A*11:01-GTS1 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
7DFG
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Li, Z, Zhou, Z, Yu, X. | | Deposit date: | 2020-11-08 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
To Be Published
|
|
7DFH
 
 | |
8P01
 
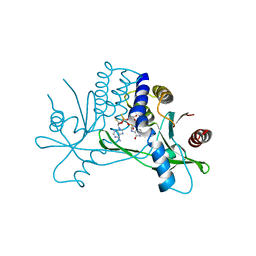 | | Crystal structure of human STING ectodomain in complex with BI 7446, a potent cyclic dinucleotide STING agonist with broad-spectrum variant activity for the treatment of cancer | | Descriptor: | 3-[(1~{R},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9-fluoranyl-18-oxidanyl-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-6~{H}-imidazo[4,5-d]pyridazin-7-one, Stimulator of interferon genes protein | | Authors: | Nar, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Discovery of BI 7446: A Potent Cyclic Dinucleotide STING Agonist with Broad-Spectrum Variant Activity for the Treatment of Cancer.
J.Med.Chem., 66, 2023
|
|
7DOK
 
 | |
7DOI
 
 | |
4F9N
 
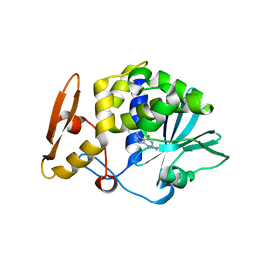 | | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with N7-methylated guanine at 2.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-7-methyl-1,7-dihydro-6H-purin-6-one, Ribosome inactivating protein | | Authors: | Yamini, S, Kushwaha, G.S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-05-19 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with N7-methylated guanine at 2.65 A resolution
To be Published
|
|
4ZX6
 
 | |
4ZY2
 
 | | X-ray crystal structure of PfA-M17 in complex with hydroxamic acid-based inhibitor 10o | | Descriptor: | CARBONATE ION, DIMETHYL SULFOXIDE, N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluorobiphenyl-4-yl)ethyl]-2,2-dimethylpropanamide, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2015-05-21 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent dual inhibitors of Plasmodium falciparum M1 and M17 aminopeptidases through optimization of S1 pocket interactions.
Eur.J.Med.Chem., 110, 2016
|
|
5WJN
 
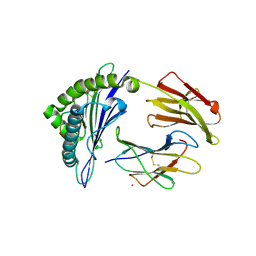 | | Crystal Structure of HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, GTS3 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
7RXS
 
 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXR
 
 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
