1RYV
 
 | |
1NIY
 
 | |
5FB1
 
 | | Crystal Structure of a PHD finger bound to histone H3 K9me3 peptide | | Descriptor: | MALONATE ION, Nuclear autoantigen Sp-100, Peptide from Histone H3, ... | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
5FB0
 
 | | Crystal Structure of a PHD finger bound to histone H3 T3ph peptide | | Descriptor: | Nuclear autoantigen Sp-100, Peptide from Histone H3, ZINC ION | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
5UJJ
 
 | |
4LSW
 
 | |
4MH1
 
 | |
9IND
 
 | | Integrin alpha-v beta-8 in complex with the Fab of 130H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-V heavy chain, Integrin beta-8, ... | | Authors: | Sun, H, Zheng, Q, Gui, X. | | Deposit date: | 2024-07-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Specifically blocking alpha v beta 8-mediated TGF-beta signaling to reverse immunosuppression by modulating macrophage polarization.
J Exp Clin Cancer Res, 44, 2025
|
|
5UJI
 
 | |
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
6BRB
 
 | | Novel non-antibody protein scaffold targeting CD40L | | Descriptor: | CD40 ligand, Tn3-like, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Baca, M, Thisted, T, Grinberg, L, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A CD40L-targeting protein reduces autoantibodies and improves disease activity in patients with autoimmunity.
Sci Transl Med, 11, 2019
|
|
7M53
 
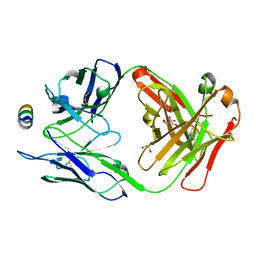 | | B6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M51
 
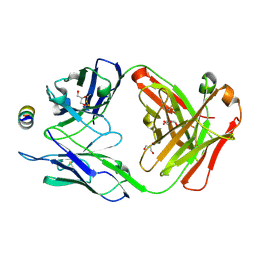 | | B6 Fab fragment bound to the OC43 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M55
 
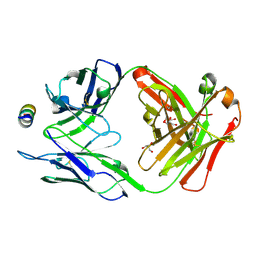 | | B6 Fab fragment bound to the MERS-CoV spike stem helix peptide | | Descriptor: | B6 antigen binding fragment (Fab) heavy chain, B6 antigen binding fragment (Fab) light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M5E
 
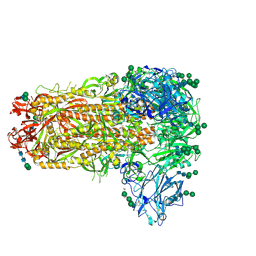 | |
7M52
 
 | | B6 Fab fragment bound to the HKU4 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
8HLC
 
 | | S protein of SARS-CoV-2 in complex with 3711 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
8HLD
 
 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Defining the features and structure of neutralizing antibody targeting the silent face of the SARS-CoV-2 spike N-terminal domain.
MedComm (2020), 5, 2024
|
|
8K3K
 
 | |
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
