8G54
 
 | |
8G58
 
 | |
6ABT
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
5XEF
 
 | | Crystal structure of flagellar chaperone from bacteria | | Descriptor: | Flagellar protein fliS | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the flagellar chaperone FliS from Bacillus cereus and an invariant proline critical for FliS dimerization and flagellin recognition
Biochem. Biophys. Res. Commun., 487, 2017
|
|
6ABQ
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
4OK7
 
 | | Structure of bacteriophage SPN1S endolysin from Salmonella typhimurium | | Descriptor: | Endolysin, GLYCEROL, SULFATE ION | | Authors: | Park, Y, Lim, J, Kong, M, Ryu, S, Rhee, S. | | Deposit date: | 2014-01-22 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of bacteriophage SPN1S endolysin reveals an unusual two-module fold for the peptidoglycan lytic and binding activity.
Mol.Microbiol., 92, 2014
|
|
6M53
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, GLYCEROL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
8YPU
 
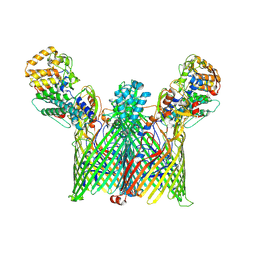 | | Cryo-EM structure of ButCD complex | | Descriptor: | Membrane protein, SusC/RagA family TonB-linked outer membrane protein | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.
Nat Commun, 15, 2024
|
|
8YPT
 
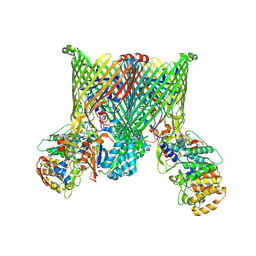 | | Cryo-EM structure of BfUbb-ButCD complex | | Descriptor: | Membrane protein, TonB-linked outer membrane protein, Ubiquitin-like protein | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.
Nat Commun, 15, 2024
|
|
4M4Y
 
 | |
5WG1
 
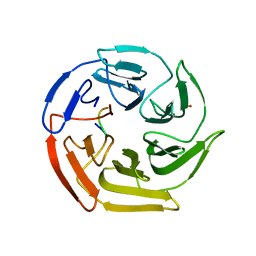 | | Kelch domain of human Keap1 bound to mutant Nrf2 EAGE peptide | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 EAGE mutant peptide, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5WFV
 
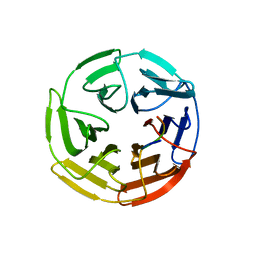 | | Kelch domain of human Keap1 bound to Nrf2 ETGE peptide | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 ETGE peptide, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5WHO
 
 | |
5WFL
 
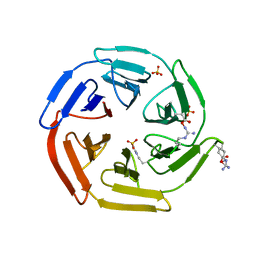 | | Kelch domain of human Keap1 in open unliganded conformation | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
6WI9
 
 | | Human secretin receptor Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Belousoff, M.J, Khoshouei, M. | | Deposit date: | 2020-04-09 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and dynamics of the active Gs-coupled human secretin receptor.
Nat Commun, 11, 2020
|
|
6WZG
 
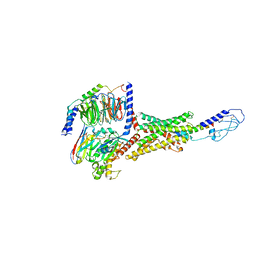 | | Human secretin receptor Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Belousoff, M.J, Danev, R. | | Deposit date: | 2020-05-13 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure and dynamics of the active Gs-coupled human secretin receptor.
Nat Commun, 11, 2020
|
|
7BPC
 
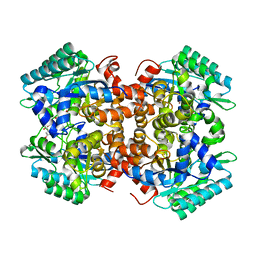 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-22 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7BP1
 
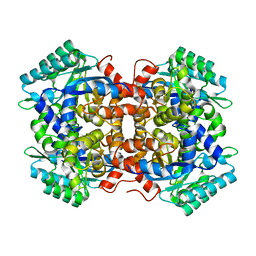 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7D85
 
 | |
4M5Y
 
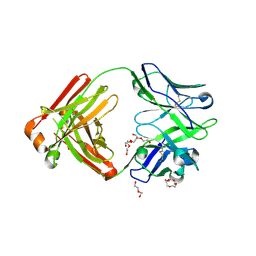 | | Crystal structure of broadly neutralizing Fab 5J8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab 5J8 heavy chain, Fab 5J8 light chain, ... | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antibody Recognition of the Pandemic H1N1 Influenza Virus Hemagglutinin Receptor Binding Site.
J.Virol., 87, 2013
|
|
5WHL
 
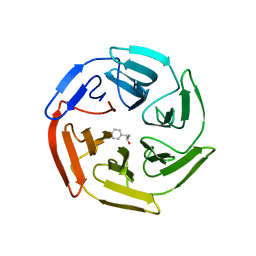 | |
5WIY
 
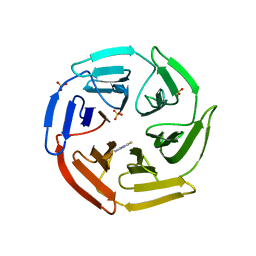 | | Kelch domain of human Keap1 bound to small molecule inhibitor fragment: 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
2NDI
 
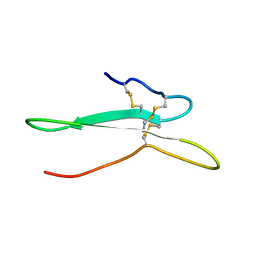 | | Solution structure of the toxin ISTX-I from Ixodes scapularis | | Descriptor: | Putative secreted salivary protein | | Authors: | Hu, K. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A sodium channel inhibitor ISTX-I with a novel structure provides a new hint at the evolutionary link between two toxin folds.
Sci Rep, 6, 2016
|
|
2OHU
 
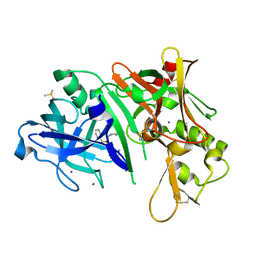 | | X-ray crystal structure of beta secretase complexed with compound 8b | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHR
 
 | | X-ray crystal structure of beta secretase complexed with compound 6a | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
