7KMR
 
 | |
3SJL
 
 | |
3SLE
 
 | |
6CI7
 
 | |
6CIB
 
 | |
8SPP
 
 | |
8TLX
 
 | |
8TLV
 
 | |
8TLW
 
 | |
3RLM
 
 | |
3RMZ
 
 | |
3PLY
 
 | | Structure of Oxidized P96G Mutant of Amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2010-11-15 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline 96 of the copper ligand loop of amicyanin regulates electron transfer from methylamine dehydrogenase by positioning other residues at the protein-protein interface.
Biochemistry, 50, 2011
|
|
3RN0
 
 | |
3SVW
 
 | |
9ILS
 
 | |
9ILR
 
 | | The structure of the GmvT-GmvA-AcCoA ternary complex | | Descriptor: | ACETYL COENZYME *A, DUF1778 domain-containing protein, N-acetyltransferase, ... | | Authors: | Xie, W, Chen, R, Zhao, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the Shigella flexneri GmvAT toxin-antitoxin system.
Febs Lett., 599, 2025
|
|
4OJK
 
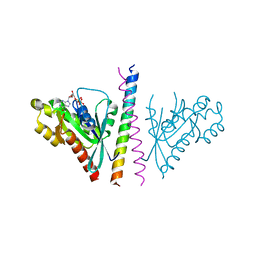 | | Structure of the cGMP Dependent Protein Kinase II and Rab11b Complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-11B, cGMP-dependent protein kinase 2 | | Authors: | Reger, A.S, Yang, M.P, Guo, E, Kim, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Crystal Structure of the cGMP-dependent Protein Kinase II Leucine Zipper and Rab11b Protein Complex Reveals Molecular Details of G-kinase-specific Interactions.
J.Biol.Chem., 289, 2014
|
|
8X2J
 
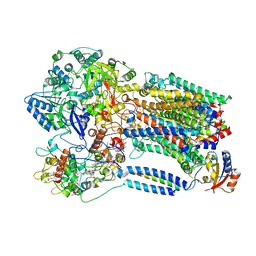 | |
2KX4
 
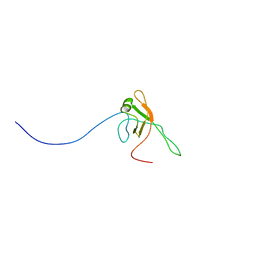 | | Solution structure of Bacteriophage Lambda gpFII | | Descriptor: | Tail attachment protein | | Authors: | Maxwell, K.L, Cardarelli, L, Neudecker, P, Davidson, A.R, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-04-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Phages have adapted the same protein fold to fulfill multiple functions in virion assembly.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7YKD
 
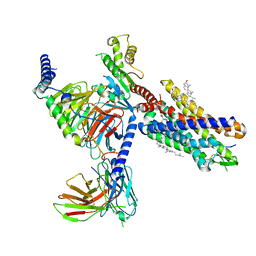 | | Cryo-EM structure of the human chemerin receptor 1 complex with the C-terminal nonapeptide of chemerin | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, G, Liao, Q, Ye, R.D, Wang, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of the human chemerin receptor 1-Gi protein complex bound to the C-terminal nonapeptide of chemerin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8K9E
 
 | |
8K9F
 
 | |
8JEI
 
 | | Cryo-EM Structure of the compuond 5c-HCAR3-Gi complex | | Descriptor: | 3-nitro-4-(propylamino)benzoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fang, Y, Pan, X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3.
Cell Rep, 43, 2024
|
|
8JEF
 
 | | Cryo-EM Structure of the 3HO-HCAR3-Gi complex | | Descriptor: | (3S)-3-hydroxyoctanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fang, Y, Pan, X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3.
Cell Rep, 43, 2024
|
|
7FJI
 
 | | human Pol III elongation complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Hou, H, Xu, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into RNA polymerase III-mediated transcription termination through trapping poly-deoxythymidine.
Nat Commun, 12, 2021
|
|
