4A72
 
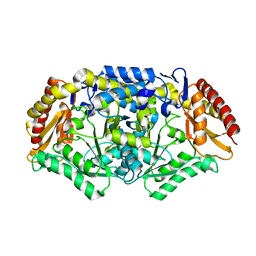 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in a mixture of apo and PLP-bound states | | Descriptor: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-10 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A6T
 
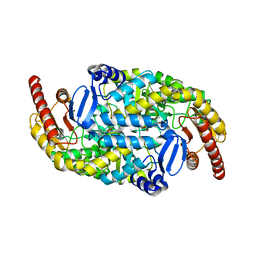 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PLP | | Descriptor: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
3WZN
 
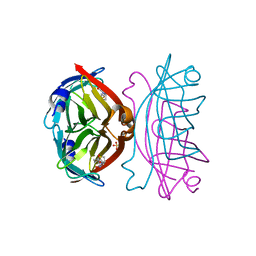 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin at 1.3 A resolution | | Descriptor: | BIOTIN, SULFATE ION, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WY8
 
 | | Crystal Structure of Protease Anisep from Arthrobacter Nicotinovorans | | Descriptor: | Serine protease | | Authors: | Sone, T, Haraguchi, Y, Kuwahara, A, Ose, T, Takano, M, Abe, A, Tanaka, M, Tanaka, I, Asano, K. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease.
Protein Pept.Lett., 22, 2015
|
|
3WZP
 
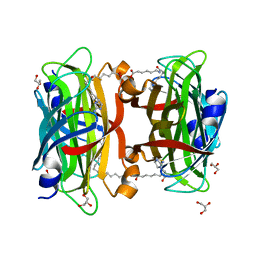 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZQ
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.7 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, HEXAETHYLENE GLYCOL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
2E6X
 
 | | X-ray structure of TT1592 from Thermus thermophilus HB8 | | Descriptor: | Hypothetical protein TTHA1281 | | Authors: | Yamada, M, Nakagawa, N, Kanagawa, M, Kuramitsu, S, Kamitori, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-05 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of TT1592 from Thermus thermophilus HB8
To be Published
|
|
2FAW
 
 | | crystal structure of papaya glutaminyl cyclase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ZINC ION, ... | | Authors: | Wintjens, R, Belrhali, H, Clantin, B, Azarkan, M, Bompard, C, Baeyens-Volant, D, Looze, Y, Villeret, V. | | Deposit date: | 2005-12-08 | | Release date: | 2006-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of papaya glutaminyl cyclase, an archetype for plant and bacterial glutaminyl cyclases.
J.Mol.Biol., 357, 2006
|
|
1WDY
 
 | | Crystal structure of ribonuclease | | Descriptor: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Goto, Y, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-05-19 | | Release date: | 2004-10-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of 2',5'-linked oligoadenylates by human ribonuclease L
Embo J., 23, 2004
|
|
1WIY
 
 | | Crystal Structure Analysis of a 6-coordinated Cytochorome P450 from Thermus thermophilus HB8 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kousumi, Y, Nakagawa, N, Kaneko, M, Yamamoto, H, Masui, R, Kuramitsu, S, Ueyama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Cytochrome P450 from Thermus thermophilus HB8
To be Published
|
|
8GAT
 
 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAV
 
 | | Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab NDS.3, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAU
 
 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
2IP4
 
 | | Crystal Structure of Glycinamide Ribonucleotide Synthetase from Thermus thermophilus HB8 | | Descriptor: | Phosphoribosylamine--glycine ligase, SULFATE ION | | Authors: | Sampei, G, Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kawai, H, Fukai, Y, Ebihara, A, Nakagawa, N, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-11 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2GS9
 
 | | Crystal structure of TT1324 from Thermus thermophilis HB8 | | Descriptor: | FORMIC ACID, Hypothetical protein TT1324, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kamitori, S, Abe, A, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-25 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of TT1324 from Thermus thermophilis HB8
To be Published
|
|
2QZR
 
 | | tRNA-Guanine Transglycosylase(TGT) in Complex with 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hoertner, S.R, Ritschel, T, Stengl, B, Kramer, C, Schweizer, W.B, Wagner, B, Kansy, M, Klebe, G, Diederich, F. | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent Inhibitors of tRNA-Guanine Transglycosylase, an Enzyme Linked to the Pathogenicity of the Shigella Bacterium: Charge-Assisted Hydrogen Bonding.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
7E5P
 
 | | Aptamer enhancing peroxidase activity of myoglobin | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*TP*TP*GP*GP*GP*AP*GP*GP*G)-3') | | Authors: | Tsukakoshi, K, Matsugami, A, Khunathai, K, Kanazashi, M, Yamagishi, Y, Nakama, K, Oshikawa, D, Hayashi, F, Kuno, H, Ikebukuro, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-quadruplex-forming aptamer enhances the peroxidase activity of myoglobin against luminol.
Nucleic Acids Res., 49, 2021
|
|
1SEK
 
 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | Descriptor: | SERPIN K | | Authors: | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
7F1X
 
 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
1V8B
 
 | | Crystal structure of a hydrolase | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, adenosylhomocysteinase | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Shiraiwa, K, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-01-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of S-Adenosyl-l-Homocysteine Hydrolase from the Human Malaria Parasite Plasmodium falciparum
J.Mol.Biol., 343, 2004
|
|
6I04
 
 | | Crystal structure of Sema domain of the Met receptor in complex with FAB | | Descriptor: | Fab heavy chain, Fab light chain, Hepatocyte growth factor receptor | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I07
 
 | | Crystal structure of EpCAM in complex with scFv | | Descriptor: | Epithelial cell adhesion molecule, GLYCEROL, Single chain Fv | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HYG
 
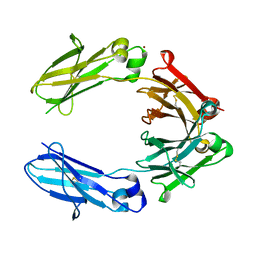 | | Heteromeric tandem IgG4/IgG1 Fc | | Descriptor: | IgHG1 and IgHG4 hybrid, ZINC ION | | Authors: | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5AJH
 
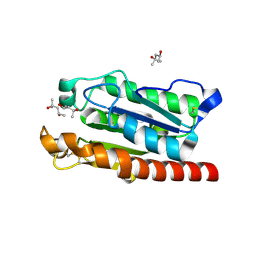 | | Crystal structure of Fusarium oxysporum cutinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CUTINASE | | Authors: | Dimarogona, M, Nikolaivits, E, Kanelli, M, Christakopoulos, P, Sandgren, M, Topakas, E. | | Deposit date: | 2015-02-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Studies of a Fusarium Oxysporum Cutinase with Polyethylene Terephthalate Modification Potential.
Biochim.Biophys.Acta, 1850, 2015
|
|
2YWC
 
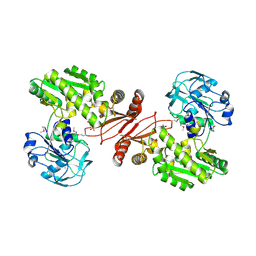 | | Crystal structure of GMP synthetase from Thermus thermophilus in complex with XMP | | Descriptor: | GMP synthase [glutamine-hydrolyzing], XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of GMP synthetase from Thermus thermophilus
To be Published
|
|
