5TRM
 
 | |
6CWS
 
 | |
5GWD
 
 | | Structure of Myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Xu, D, Ran, T, Wang, W. | | Deposit date: | 2016-09-10 | | Release date: | 2017-02-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Myroilysin Is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Is Activated by a Cysteine Switch Mechanism.
J. Biol. Chem., 292, 2017
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
5ME9
 
 | | Crystal structure of yeast Cdt1 (N terminal and middle domain), form 1. | | Descriptor: | Cell division cycle protein CDT1, GLYCEROL, SULFATE ION | | Authors: | Pye, V.E, Frigola, J, Diffley, J.F.X, Cherepanov, P. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdt1 stabilizes an open MCM ring for helicase loading.
Nat Commun, 8, 2017
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
6KKL
 
 | | Crystal structure of Drug:Proton Antiporter-1 (DHA1) Family SotB, in the inward conformation (H115N mutant) | | Descriptor: | Sugar efflux transporter, nonyl beta-D-glucopyranoside | | Authors: | Xiao, Q.J, Sun, B, Zuo, Y.X, Guo, L, He, J.H, Deng, D. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Visualizing the nonlinear changes of a drug-proton antiporter from inward-open to occluded state.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
4HXN
 
 | | Brd4 Bromodomain 1 complex with 4-(2-FLUOROPHENYL)-1,3-THIAZOL-2(3H)-ONE inhibitor | | Descriptor: | 4-(2-fluorophenyl)-1,3-thiazol-2(3H)-one, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXP
 
 | | Brd4 Bromodomain 1 complex with 4-(2-OXO-1,3-OXAZOLIDIN-3-YL)BENZAMIDE inhibitor | | Descriptor: | 4-(2-oxo-1,3-oxazolidin-3-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXS
 
 | | Brd4 Bromodomain 1 complex with N-[3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)PHENYL]-1-PHENYLMETHANESULFONAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]-1-phenylmethanesulfonamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXL
 
 | | Brd4 Bromodomain 1 complex with 3-CYCLOHEXYL-N-{3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)-5-[(THIOPHEN-2-YLSULFONYL)AMINO]PHENYL}PROPANAMIDE inhibitor | | Descriptor: | 3-cyclohexyl-N-{3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)-5-[(thiophen-2-ylsulfonyl)amino]phenyl}propanamide, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXM
 
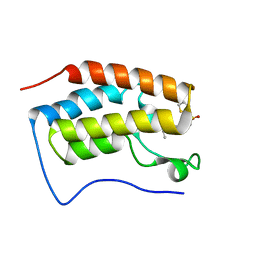 | | Brd4 Bromodomain 1 complex with N-{3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)-5-[(THIOPHEN-2-YLSULFONYL)AMINO]PHENYL}BUTANAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-{3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)-5-[(thiophen-2-ylsulfonyl)amino]phenyl}butanamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXO
 
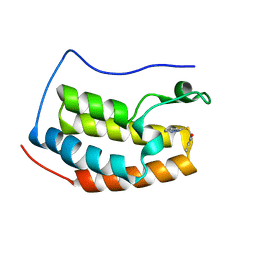 | | Brd4 Bromodomain 1 complex with 3-{[(3-METHYL-1,2-OXAZOL-5-YL)METHYL]SULFANYL}[1,2,4]TRIAZOLO[4,3-A]PYRIDINE inhibitor | | Descriptor: | 3-{[(3-methyl-1,2-oxazol-5-yl)methyl]sulfanyl}[1,2,4]triazolo[4,3-a]pyridine, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXK
 
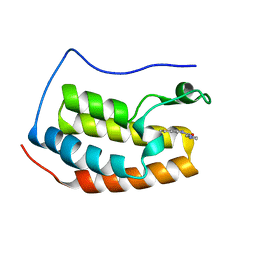 | | Brd4 Bromodomain 1 complex with 6,7-DIHYDROTHIENO[3,2-C]PYRIDIN-5(4H)-YL(1H-IMIDAZOL-1-YL)METHANONE inhibitor | | Descriptor: | 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl(1H-imidazol-1-yl)methanone, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXR
 
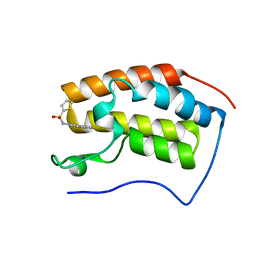 | | Brd4 Bromodomain 1 complex with N-[3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)PHENYL]THIOPHENE-2-SULFONAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]thiophene-2-sulfonamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
7XVQ
 
 | | Crystal structure of AcrIIC4 | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, H, Li, X.Z, Song, G.Y. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-09 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitory mechanism of CRISPR-Cas9 by AcrIIC4.
Nucleic Acids Res., 51, 2023
|
|
8WWP
 
 | | PNPase mutant of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
2REU
 
 | | Crystal Structure of the C-terminal of Sau3AI fragment | | Descriptor: | MAGNESIUM ION, Type II restriction enzyme Sau3AI | | Authors: | Hu, X, Yu, F, Xu, C, He, J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of C-terminal Sau3AI domain
Biochim.Biophys.Acta, 1794, 2009
|
|
5GUF
 
 | |
6JOP
 
 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | MAGNESIUM ION, Primase, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6JOQ
 
 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
8HF0
 
 | | DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state | | Descriptor: | Dicer-2, isoform A, LD06392p, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural mechanism of R2D2 and Loqs-PD synergistic modulation on DmDcr-2 oligomers.
Nat Commun, 14, 2023
|
|
8HF1
 
 | | DmDcr-2/R2D2/LoqsPD with 19bp-dsRNA in Trimer state | | Descriptor: | Dicer-2, isoform A, LD06392p, ... | | Authors: | Su, S, Wang, J, Ma, J. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of R2D2 and Loqs-PD synergistic modulation on DmDcr-2 oligomers.
Nat Commun, 14, 2023
|
|
6JON
 
 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | Authors: | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6KJU
 
 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
