3N72
 
 | | Crystal Structure of Aha-1 from plasmodium falciparum, PFC0270w | | Descriptor: | THIOCYANATE ION, putative activator of HSP90 | | Authors: | Wernimont, A.K, Dong, A, Hutchinson, A, Sullivan, H, Mackenzie, F, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of Aha-1 from plasmodium falciparum, PFC0270w
To be Published
|
|
3NIZ
 
 | | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rhodanese family protein | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Lin, Y.H, Hassanali, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound.
To be Published
|
|
8FBG
 
 | | Crystal structure of NSD1 Mutant-Y1869C | | Descriptor: | CALCIUM ION, Histone-lysine N-methyltransferase, H3 lysine-36 specific, ... | | Authors: | Providokhina, K, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NSD1
To Be Published
|
|
8FBH
 
 | | Crystal structure of NSD1 Mutant-T1927A | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Providokhina, K, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of NSD1
To Be Published
|
|
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
3OI7
 
 | | Structure of the structure of the H13A mutant of Ykr043C in complex with sedoheptulose-1,7-bisphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1,7-di-O-phosphono-beta-D-altro-hept-2-ulofuranose, GLYCEROL, ... | | Authors: | Singer, A.U, Xu, X, Dong, A, Cui, H, Clasquin, M.F, Caudy, A.A, Edwards, A.M, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Riboneogenesis in yeast.
Cell(Cambridge,Mass.), 145, 2011
|
|
8G5E
 
 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, N~4~-[6-(dimethylamino)hexyl]-N~2~-[5-(dimethylamino)pentyl]-6,7-dimethoxyquinazoline-2,4-diamine, UNKNOWN ATOM OR ION | | Authors: | Beldar, S, Dong, A, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535
To be published
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
2NYR
 
 | | Crystal Structure of Human Sirtuin Homolog 5 in Complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, NAD-dependent deacetylase sirtuin-5, ZINC ION | | Authors: | Min, J.R, Antoshenko, T, Allali-Hassani, A, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin.
Structure, 15, 2007
|
|
8G2G
 
 | | Crystal structure of PRMT3 with compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3, SULFATE ION | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-02-03 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G2F
 
 | | Crystal Structure of PRMT3 with Compound II710 | | Descriptor: | 5'-S-[3-(N'-benzylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G2H
 
 | | Crystal Structure of PRMT4 with Compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G2I
 
 | | Crystal Structure of PRMT4 with Compound YD1290 | | Descriptor: | 5'-([2-(benzylcarbamamido)ethyl]{3-[N'-(3-bromophenyl)carbamimidamido]propyl}amino)-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
7KLJ
 
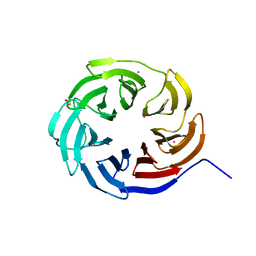 | | Crystal structure of the WD-repeat domain of human KIF21A | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of Kinesin-like protein KIF21A, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Loppnau, P, Hutchinson, A, Seitova, A, Edwards, A.M, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of the WD-repeat domain of human KIF21A
To be Published
|
|
7KYX
 
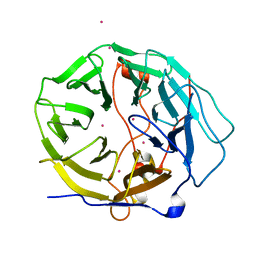 | | Crystal structure of human CORO6 | | Descriptor: | Coronin-6, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Loppnau, P, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of human CORO6
To Be Published
|
|
4XCX
 
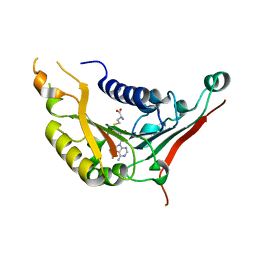 | | METHYLTRANSFERASE DOMAIN OF SMALL RNA 2'-O-METHYLTRANSFERASE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Small RNA 2'-O-methyltransferase | | Authors: | Walker, J.R, Zeng, H, Dong, A, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of human C1ORF59 in complex with SAH
To be published
|
|
4XVH
 
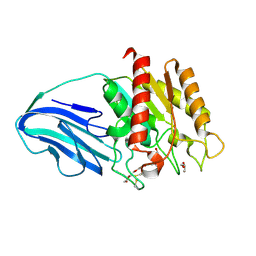 | | Crystal structure of a Corynascus thermopiles (Myceliophthora fergusii) carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD) | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Carbohydrate esterase family 2 (CE2), GLYCEROL | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-27 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9449 Å) | | Cite: | Crystal structure of a Corynascus thermopiles carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD)
To Be Published
|
|
4XUY
 
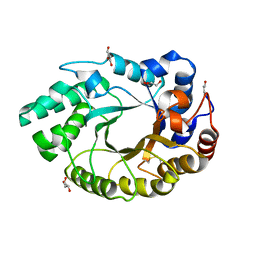 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger | | Descriptor: | GLYCEROL, Probable endo-1,4-beta-xylanase C, SULFATE ION | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0027 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger
To Be Published
|
|
7MT1
 
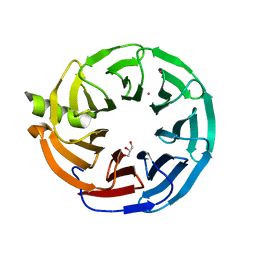 | | Crystal structure of Human Platelet-activating factor acetylhydrolase IB subunit beta (PAFAH1B1) | | Descriptor: | GLYCEROL, Platelet-activating factor acetylhydrolase IB subunit beta, UNKNOWN ATOM OR ION | | Authors: | Hutchinson, A, Seitova, A, Dong, A, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of Human Platelet-activating factor acetylhydrolase IB subunit beta (PAFAH1B1)
To Be Published
|
|
7MDN
 
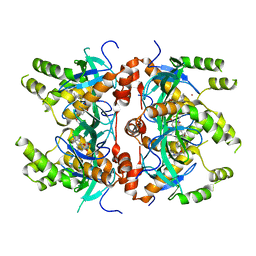 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
6BPI
 
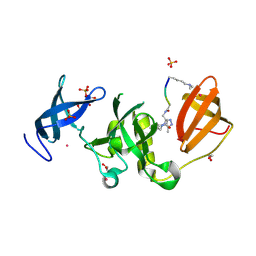 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
2I81
 
 | | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced | | Descriptor: | 2-Cys Peroxiredoxin | | Authors: | Artz, J.D, Qiu, W, Dong, A, Lew, J, Ren, H, Zhao, Y, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced
To be published
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
2QKR
 
 | | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with indirubin 3'-monoxime bound | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, Cdc2-like CDK2/CDC28 like protein kinase | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Lin, Y.H, Hassanali, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-11 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with indirubin 3'-monoxime bound.
To be Published
|
|
7M3X
 
 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
