6V91
 
 | |
6VJU
 
 | |
6W2O
 
 | |
3MBM
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with cytosine and FoL fragment 717, imidazo[2,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
5DXD
 
 | |
5DLE
 
 | |
1BL0
 
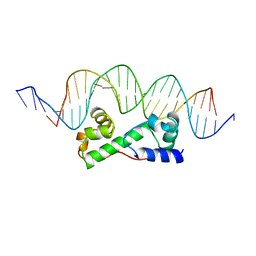 | | MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN (MARA)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP* CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP* TP*C)-3'), PROTEIN (MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN) | | Authors: | Davies, S, Rhee, R.G, Martin, J.L, Rosner, D.R. | | Deposit date: | 1998-07-22 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
6MBQ
 
 | | Crystal structure of Mg-free wild-type KRAS (2-166) bound to GMPPNP in the state 1 conformation | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Dharmaiah, S, Davies, D.R, Abendroth, J, Gillette, W.G, Stephen, A.G, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
4NI9
 
 | | Crystal structure of human interleukin 6 in complex with a modified nucleotide aptamer (SOMAMER SL1025), FORM 2 | | Descriptor: | Interleukin-6, SODIUM ION, SOMAmer SL1025 | | Authors: | Davies, D, Edwards, T, Gelinas, A, Jarvis, T, Clifton, M.C. | | Deposit date: | 2013-11-05 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of interleukin-6 in complex with a modified nucleic Acid ligand.
J.Biol.Chem., 289, 2014
|
|
4NI7
 
 | | Crystal structure of human interleukin 6 in complex with a modified nucleotide aptamer (SOMAMER SL1025) | | Descriptor: | Interleukin-6, SODIUM ION, SOMAmer SL1025 | | Authors: | Davies, D, Edwards, T, Gelinas, A, Jarvis, T, Clifton, M.C. | | Deposit date: | 2013-11-05 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of interleukin-6 in complex with a modified nucleic Acid ligand.
J.Biol.Chem., 289, 2014
|
|
3ER5
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, H-189 | | Authors: | Bailey, D, Veerapandian, B, Cooper, J, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin.
Biochem.J., 289 ( Pt 2), 1993
|
|
4ER1
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, N-[(1R,2R,4R)-1-(cyclohexylmethyl)-2-hydroxy-6-methyl-4-{[(2R)-2-methylbutyl]carbamoyl}heptyl]-3-(1H-imidazol-3-ium-4-yl)-N~2~-[3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanoyl]-L-alaninamide | | Authors: | Quail, J.W, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-14 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
4APE
 
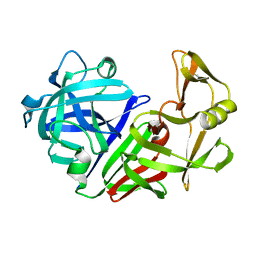 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
4ER2
 
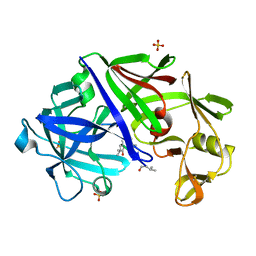 | | The active site of aspartic proteinases | | Descriptor: | ENDOTHIAPEPSIN, PEPSTATIN, SULFATE ION | | Authors: | Bailey, D, Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
4ER4
 
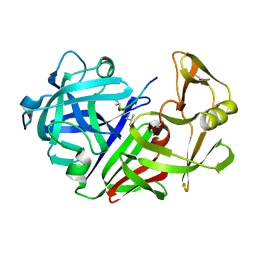 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
7K86
 
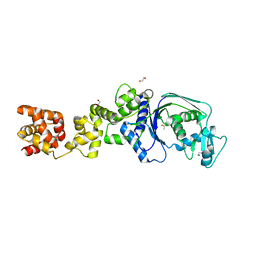 | |
7L9R
 
 | |
2ER9
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES. | | Descriptor: | ENDOTHIAPEPSIN, L363,564 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
2ER7
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES.III. THREE-DIMENSIONAL STRUCTURE OF ENDOTHIAPEPSIN COMPLEXED WITH A TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | ENDOTHIAPEPSIN, SULFATE ION, TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN | | Authors: | Veerapandian, B, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-11-12 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.
J.Mol.Biol., 216, 1990
|
|
2ER6
 
 | | The structure of a synthetic pepsin inhibitor complexed with endothiapepsin. | | Descriptor: | ENDOTHIAPEPSIN, H-256 peptide | | Authors: | Cooper, J.B, Foundling, S.I, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-13 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a synthetic pepsin inhibitor complexed with endothiapepsin
Eur.J.Biochem., 169, 1987
|
|
2ER0
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, L364,099 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
4PQ9
 
 | |
7N4D
 
 | |
4PY2
 
 | |
