4PBD
 
 | |
4PCK
 
 | | Crystal structure of the P22S mutant of N-terminal CS domain of human Shq1 | | Descriptor: | GLYCEROL, Protein SHQ1 homolog | | Authors: | Singh, M, Wang, Z, Cascio, D, Feigon, J. | | Deposit date: | 2014-04-15 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure and Interactions of the CS Domain of Human H/ACA RNP Assembly Protein Shq1.
J.Mol.Biol., 427, 2015
|
|
4GUN
 
 | |
3KX9
 
 | |
4RZT
 
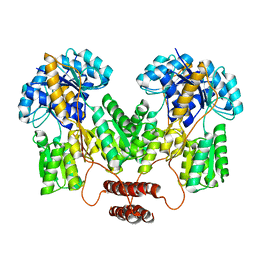 | | Lac repressor engineered to bind sucralose, sucralose-bound tetramer | | Descriptor: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Sawaya, M.R, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
4RZS
 
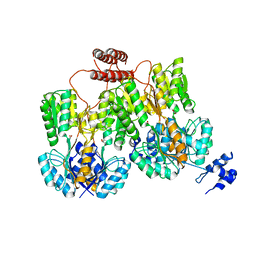 | | Lac repressor engineered to bind sucralose, unliganded tetramer | | Descriptor: | GLYCEROL, Lac repressor | | Authors: | Arbing, M.A, Cascio, D, Kosuri, S, Church, G.M. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
6BWZ
 
 | | SYSGYS from low-complexity domain of FUS, residues 37-42 | | Descriptor: | SYSGYS peptide from low-complexity domain of FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Tamir, G, Eisenberg, D.S. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
3G3V
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
6BZP
 
 | | STGGYG from low-complexity domain of FUS, residues 77-82 | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, RNA-binding protein FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
3G3W
 
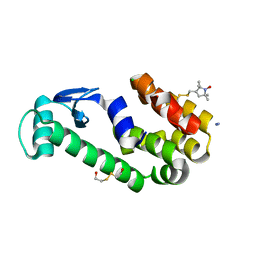 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
6DGB
 
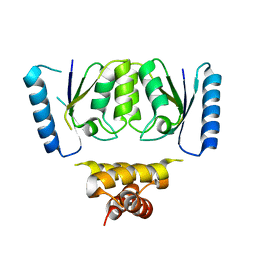 | | Crystal structure of the C-terminal catalytic domain of IS1535 TnpA, an IS607-like serine recombinase | | Descriptor: | IS607 family transposase IS1535 | | Authors: | Chen, W.Y, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
3N79
 
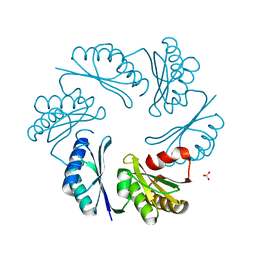 | | PduT C38S Mutant from Salmonella enterica Typhimurium | | Descriptor: | CHLORIDE ION, PduT, SODIUM ION, ... | | Authors: | Crowley, C.S, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into the Mechanisms of Transport across the Salmonella enterica Pdu Microcompartment Shell.
J.Biol.Chem., 285, 2010
|
|
6BZM
 
 | | GFGNFGTS from low-complexity/FG repeat domain of Nup98, residues 116-123 | | Descriptor: | Nuclear pore complex protein Nup98-Nup96 | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Chong, L, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-24 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
3NG0
 
 | |
6DGC
 
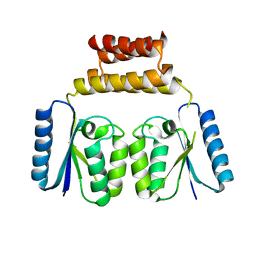 | | Crystal structure of the C-terminal catalytic domain of ISC1926 TnpA, an IS607-like serine recombinase | | Descriptor: | ISC1926 TnpA C-terminal catalytic domain | | Authors: | Hancock, S.P, Kumar, P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
6DCB
 
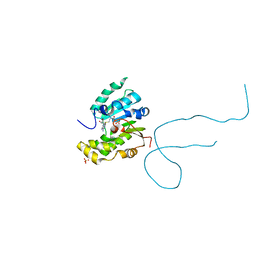 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
6BXV
 
 | | SYSSYGQS from low-complexity domain of FUS, residues 54-61 | | Descriptor: | FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6DCC
 
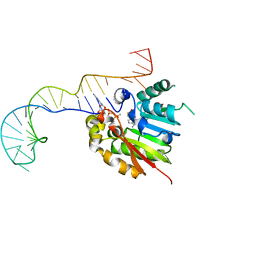 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
6BXX
 
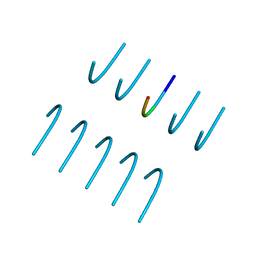 | | GYNGFG from low-complexity domain of hnRNPA1, residues 243-248 | | Descriptor: | hnRNPA1 | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6DKQ
 
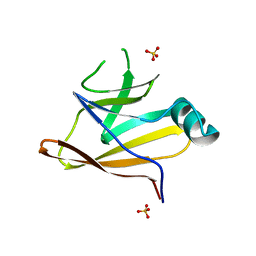 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 | | Descriptor: | Heme-binding protein Shr, SULFATE ION | | Authors: | Macdonald, R, Cascio, D, Collazo, M.J, Clubb, R.T. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Streptococcus pyogenes Shr protein captures human hemoglobin using two structurally unique binding domains.
J.Biol.Chem., 293, 2018
|
|
4QX1
 
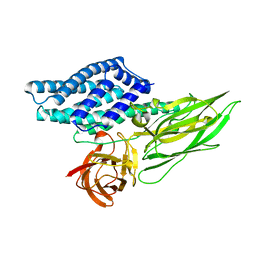 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RF2
 
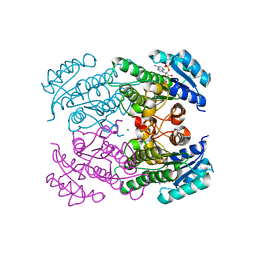 | |
4RF4
 
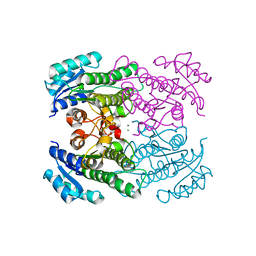 | |
4RKP
 
 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (apo form) | | Descriptor: | ACETATE ION, Putative uncharacterized protein Ta1305, SULFATE ION | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RKZ
 
 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate 3-Phosphate/ADP Bound) | | Descriptor: | (3R)-5-hydroxy-3-methyl-3-(phosphonooxy)pentanoic acid, ADENOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein Ta1305, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
