4O1B
 
 | | The crystal structure of a mutant NAMPT (G217R) in complex with an inhibitor APO866 | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1D
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
5WOP
 
 | | High Resolution Structure of Mutant CA09-PB2cap | | Descriptor: | GLYCEROL, Polymerase PB2 | | Authors: | Constantinides, A.E, Gumpper, R.H, Severin, C, Luo, M. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution structure of the Influenza A virus PB2cap binding domain illuminates the changes induced by ligand binding.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6KDR
 
 | |
6KDS
 
 | |
6KDQ
 
 | |
6LJM
 
 | |
6LJN
 
 | |
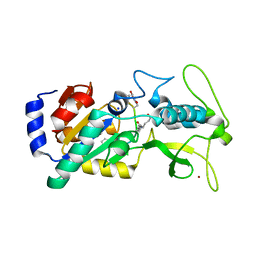
6LJK
 
 | |
8HN9
 
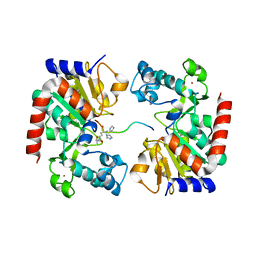 | | Human SIRT3 Recognizing CCNE2K348la peptide | | Descriptor: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Wang, Y, Ding, W. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
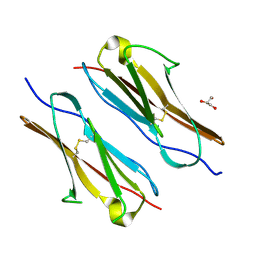
7Y73
 
 | |
7Y8I
 
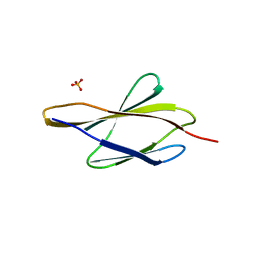 | | Crystal structure of sDscam FNIII3 domain, isoform alpha7 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dscam, ... | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y9A
 
 | | Crystal structure of sDscam Ig1-2 domains, isoform beta2v6 | | Descriptor: | Down Syndrome Cell Adhesion Molecules, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)][beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y6O
 
 | |
7Y4X
 
 | |
7Y5J
 
 | |
7Y8S
 
 | |
7Y95
 
 | | Crystal structure of sDscam Ig1 domain, isoform beta6v2 | | Descriptor: | Dscam, GLYCEROL, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y54
 
 | |
7Y5R
 
 | |
7Y6E
 
 | |
7Y8H
 
 | |
7FAQ
 
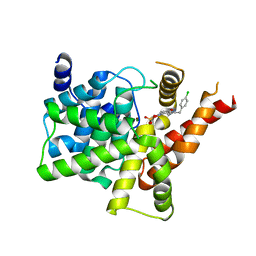 | | Crystal structure of PDE5A in complex with inhibitor L1 | | Descriptor: | 2-[(4-chlorophenyl)methyl]-5-methoxy-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.200136 Å) | | Cite: | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
7FAR
 
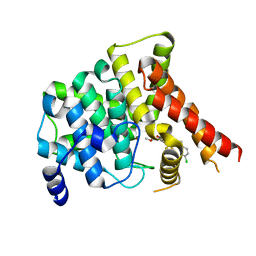 | | Crystal structure of PDE5A in complex with inhibitor L12 | | Descriptor: | 5-[bis(fluoranyl)methoxy]-2-[(4-chlorophenyl)methyl]-10-(3-methoxypropyl)-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40006471 Å) | | Cite: | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
7VS2
 
 | | secreted fungal effector protein MoErs1 | | Descriptor: | MoErs1 | | Authors: | Wang, F.F, Xing, W.M. | | Deposit date: | 2021-10-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Magnaporthe oryzae effector MoErs1 and host papain-like protease OsRD21 interaction to combat rice blast.
Nat.Plants, 2024
|
|
