6OP0
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (R)-{1-[(2,5-dimethylphenyl)methyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fairman, J.W, Davies, D.R, Foley, A, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
4ER4
 
 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
4ER1
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, N-[(1R,2R,4R)-1-(cyclohexylmethyl)-2-hydroxy-6-methyl-4-{[(2R)-2-methylbutyl]carbamoyl}heptyl]-3-(1H-imidazol-3-ium-4-y l)-N~2~-[3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanoyl]-L-alaninamide | | Authors: | Quail, J.W, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-14 | | Release date: | 1991-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
4ER2
 
 | | The active site of aspartic proteinases | | Descriptor: | ENDOTHIAPEPSIN, PEPSTATIN, SULFATE ION | | Authors: | Bailey, D, Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
3D6B
 
 | |
1IKQ
 
 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKP
 
 | | Pseudomonas Aeruginosa Exotoxin A, P201Q, W281A mutant | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
3ER5
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, H-189 | | Authors: | Bailey, D, Veerapandian, B, Cooper, J, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin.
Biochem.J., 289 ( Pt 2), 1993
|
|
3ER3
 
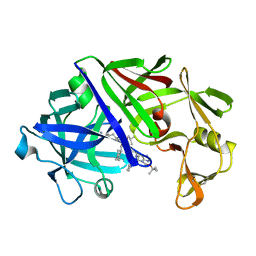 | | The active site of aspartic proteinases | | Descriptor: | 6-ammonio-N-[(2R,4R,5R)-5-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-6-cyclohexyl-4-hydroxy-2-(2-methylpropyl)hexanoyl]-L-norleucylphenylalanine, ENDOTHIAPEPSIN | | Authors: | Al-Karadaghi, S, Cooper, J.B, Veerapandian, B, Hoover, D, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
6VC8
 
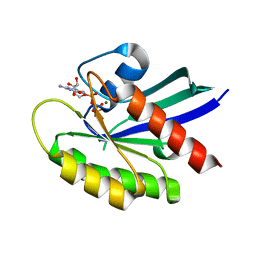 | | Crystal structure of wild-type KRAS4b(1-169) in complex with GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Davies, D.R, Edwards, T.E, Simanshu, D.K. | | Deposit date: | 2019-12-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6NUP
 
 | |
3P0Z
 
 | |
3MBM
 
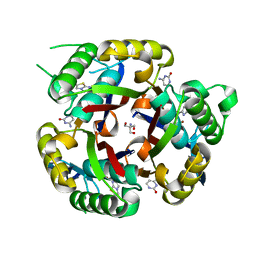 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with cytosine and FoL fragment 717, imidazo[2,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
3P08
 
 | |
3P10
 
 | |
3RQI
 
 | |
3QHD
 
 | |
7T7J
 
 | |
7U5Y
 
 | |
7U5Q
 
 | |
7SY9
 
 | |
2ER9
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES. | | Descriptor: | ENDOTHIAPEPSIN, L363,564 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
2ER7
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES.III. THREE-DIMENSIONAL STRUCTURE OF ENDOTHIAPEPSIN COMPLEXED WITH A TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | ENDOTHIAPEPSIN, SULFATE ION, TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN | | Authors: | Veerapandian, B, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-11-12 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.
J.Mol.Biol., 216, 1990
|
|
2ER6
 
 | | The structure of a synthetic pepsin inhibitor complexed with endothiapepsin. | | Descriptor: | ENDOTHIAPEPSIN, H-256 peptide | | Authors: | Cooper, J.B, Foundling, S.I, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-13 | | Release date: | 1991-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a synthetic pepsin inhibitor complexed with endothiapepsin
Eur.J.Biochem., 169, 1987
|
|
2ER0
 
 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, L364,099 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
