5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
4V47
 
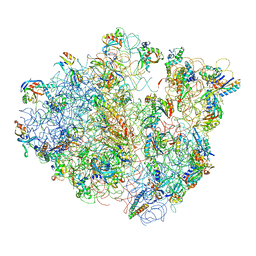 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
5FLZ
 
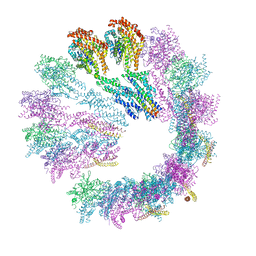 | | Cryo-EM structure of gamma-TuSC oligomers in a closed conformation | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
7M3P
 
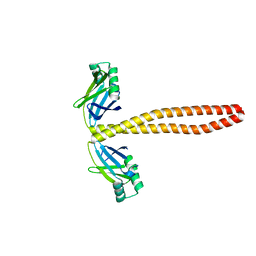 | | Xrcc4-Spc110p(164-207) fusion | | Descriptor: | Xrcc4-Spc110p(164-207) | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0000186 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2Y
 
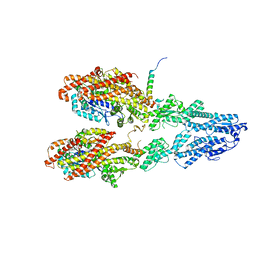 | | Closed conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2X
 
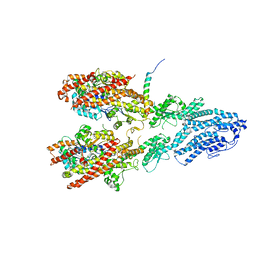 | | Open conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2W
 
 | | Engineered disulfide cross-linked closed conformation of the Yeast gamma-TuRC(SS) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2Z
 
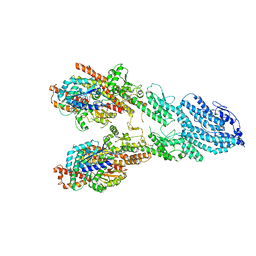 | | Monomeric single-particle reconstruction of the Yeast gamma-TuSC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component SPC97, Spindle pole body component SPC98, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
7SP5
 
 | | Crystal Structure of a Eukaryotic Phosphate Transporter | | Descriptor: | PHOSPHATE ION, Phosphate transporter, nonyl beta-D-glucopyranoside | | Authors: | Stroud, R.M, Pedersen, B.P, Kumar, H, Waight, A.B, Risenmay, A.J, Roe-Zurz, Z, Chau, B.H, Schlessinger, A, Bonomi, M, Harries, W, Sali, A, Johri, A.K, Finer-Moore, J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a eukaryotic phosphate transporter.
Nature, 496, 2013
|
|
6DRD
 
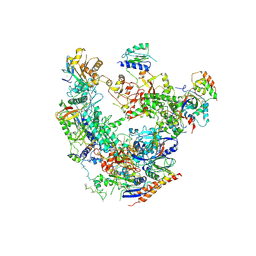 | | RNA Pol II(G) | | Descriptor: | DNA-directed RNA polymerase II subunit GRINL1A, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Yu, X, Jishage, M, Shi, Y, Ganesan, S, Sali, A, Chait, B.T, Asturias, F, Roeder, R.G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-12 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5FM1
 
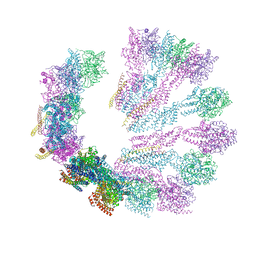 | | Structure of gamma-tubulin small complex based on a cryo-EM map, chemical cross-links, and a remotely related structure | | Descriptor: | SPINDLE POLE BODY COMPONENT 110, SPINDLE POLE BODY COMPONENT SPC97, SPINDLE POLE BODY COMPONENT SPC98, ... | | Authors: | Greenberg, C.H, Kollman, J, Zelter, A, Johnson, R, MacCoss, M.J, Davis, T.N, Agard, D.A, Sali, A. | | Deposit date: | 2015-10-30 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
J.Struct.Biol., 194, 2016
|
|
1I9A
 
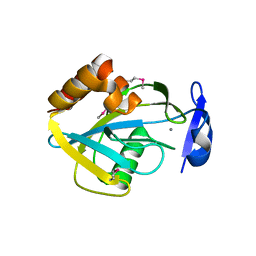 | | STRUCTURAL STUDIES OF CHOLESTEROL BIOSYNTHESIS: MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AND ISOPENTENYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-03-18 | | Release date: | 2001-03-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3NF5
 
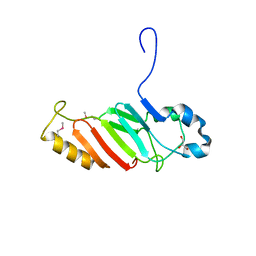 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
1BO4
 
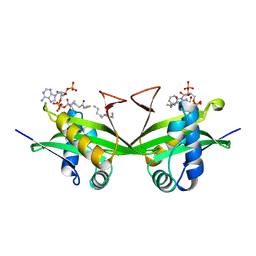 | | CRYSTAL STRUCTURE OF A GCN5-RELATED N-ACETYLTRANSFERASE: SERRATIA MARESCENS AMINOGLYCOSIDE 3-N-ACETYLTRANSFERASE | | Descriptor: | COENZYME A, PROTEIN (SERRATIA MARCESCENS AMINOGLYCOSIDE-3-N-ACETYLTRANSFERASE), SPERMIDINE | | Authors: | Wolf, E, Vassilev, A, Makino, Y, Sali, A, Nakatani, Y, Burley, S.K. | | Deposit date: | 1998-08-08 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase.
Cell(Cambridge,Mass.), 94, 1998
|
|
4V48
 
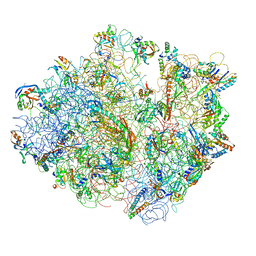 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
3B63
 
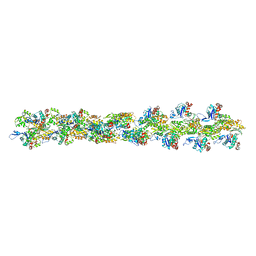 | | Actin filament model in the extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments.
J.Mol.Biol., 375, 2008
|
|
3B5U
 
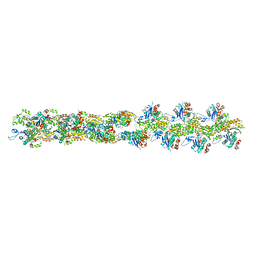 | | Actin filament model from extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments
J.Mol.Biol., 375, 2008
|
|
6XMJ
 
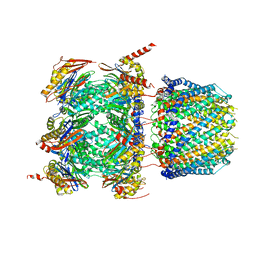 | | Human 20S proteasome bound to an engineered 11S (PA26) activator | | Descriptor: | Proteasome activator protein PA26, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | de la Pena, A.H, Opoku-Nsiah, K.A, Williams, S.K, Chopra, N, Sali, A, Gestwicki, J.E, Lander, G.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Y Phi motif defines the structure-activity relationships of human 20S proteasome activators.
Nat Commun, 13, 2022
|
|
4V7H
 
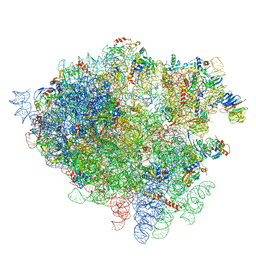 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
1A5T
 
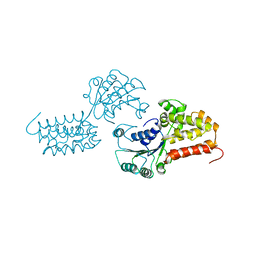 | | CRYSTAL STRUCTURE OF THE DELTA PRIME SUBUNIT OF THE CLAMP-LOADER COMPLEX OF ESCHERICHIA COLI DNA POLYMERASE III | | Descriptor: | DELTA PRIME, ZINC ION | | Authors: | Guenther, B, Onrust, R, Sali, A, O'Donnell, M, Kuriyan, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the delta' subunit of the clamp-loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 91, 1997
|
|
1CMO
 
 | | IMMUNOGLOBULIN MOTIF DNA-RECOGNITION AND HETERODIMERIZATION FOR THE PEBP2/CBF RUNT-DOMAIN | | Descriptor: | POLYOMAVIRUS ENHANCER BINDING PROTEIN 2 | | Authors: | Nagata, T, Gupta, V, Sorce, D, Kim, W.Y, Sali, A, Chait, B.T, Shigesada, K, Ito, Y, Werner, M.H. | | Deposit date: | 1999-05-11 | | Release date: | 2000-01-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain.
Nat.Struct.Biol., 6, 1999
|
|
2GDQ
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution | | Descriptor: | yitF | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-16 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution
To be Published
|
|
