3KU4
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
6UIN
 
 | | Role of Beta-hairpin motifs in the DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*NP*NP*NP*NP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | Tethering-facilitated DNA 'opening' and complementary roles of beta-hairpin motifs in the Rad4/XPC DNA damage sensor protein
Nucleic Acids Res., 48, 2021
|
|
6UG1
 
 | | Sequence impact in DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Impact of DNA sequences on DNA 'opening' by the Rad4/XPC nucleotide excision repair complex.
DNA Repair (Amst), 107, 2021
|
|
3KUK
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2'-DEOXYURIDINE, SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KVY
 
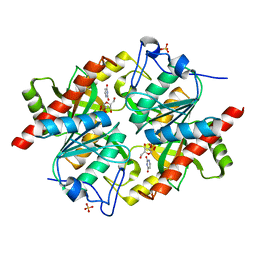 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, SULFATE ION, URACIL, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KVR
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2,5-anhydro-4-deoxy-D-erythro-pent-4-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3NL5
 
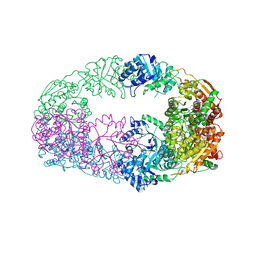 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3NM1
 
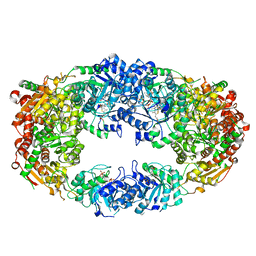 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3NL2
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | Thiamine biosynthetic bifunctional enzyme | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3NL6
 
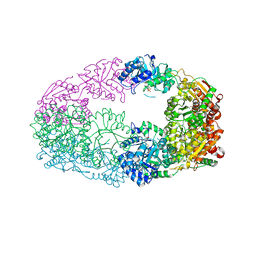 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, THIAMIN PHOSPHATE, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3NL3
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | MAGNESIUM ION, THIAMIN PHOSPHATE, Thiamine biosynthetic bifunctional enzyme | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3NM3
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3KVV
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
4MEJ
 
 | | Crystal structure of Lactobacillus helveticus purine deoxyribosyl transferase (PDT) with the tricyclic purine 8,9-dihydro-9-oxoimidazo[2,1-b]purine (N2,3-ethenoguanine) | | Descriptor: | 3H-imidazo[2,1-b]purin-4(5H)-one, Nucleoside deoxyribosyltransferase, SULFATE ION | | Authors: | Paul, D, Seckute, J, Ealick, S.E. | | Deposit date: | 2013-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosylation of a tricyclic purine analog at alternative
sites by nucleoside 2 -deoxyribosyltransferases
Plos One, 2014
|
|
5KKN
 
 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | Descriptor: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | Authors: | Wang, R, Paul, D, Tong, L. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3L8P
 
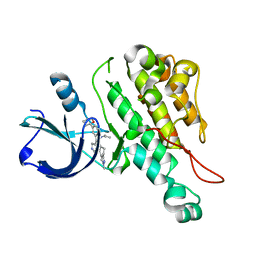 | | Crystal structure of cytoplasmic kinase domain of Tie2 complexed with inhibitor CEP11207 | | Descriptor: | 2-methyl-11-(1-methylethyl)-8-[(2S)-tetrahydro-2H-pyran-2-yl]-2,11,12,13-tetrahydro-4H-indazolo[5,4-a]pyrrolo[3,4-c]carbazol-4-one, Angiopoietin-1 receptor | | Authors: | Fedorov, A.A, Fedorov, E.V, Pauletti, D, Meyer, S.L, Hudkins, R.L, Almo, S.C. | | Deposit date: | 2010-01-03 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of cytoplasmic kinase domain of Tie2 complexed with inhibitor CEP11207
To be Published
|
|
6UBF
 
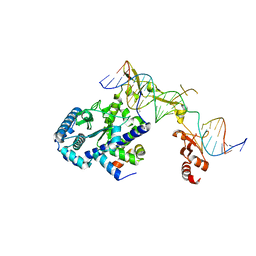 | |
6CFI
 
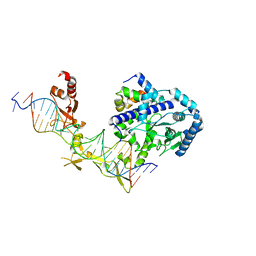 | | Crystal structure of Rad4-Rad23 bound to a 6-4 photoproduct UV lesion | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*(T64)P*TP*GP*GP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), DNA repair protein RAD4, DNA('-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*AP*AP*AP*GP*CP*TP*AP*CP*AP*A)-'), ... | | Authors: | Min, J, Jeffrey, P.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.36241913 Å) | | Cite: | Structure and mechanism of pyrimidine-pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex.
Nucleic Acids Res., 47, 2019
|
|
2LVG
 
 | |
6O4Y
 
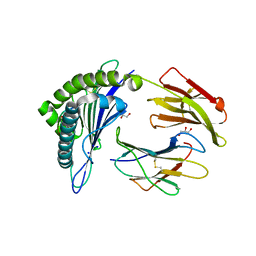 | | Structure of HLA-A2:01 with peptide MM91 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O53
 
 | | Structure of HLA-A2:01 with peptide MM96 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O4Z
 
 | | Structure of HLA-A2:01 with peptide MM92 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O51
 
 | | Structure of HLA-A2:01 with peptide MM90 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Ying, G, Bitra, A, Zajonc, D.M. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
1MA7
 
 | |
