1JP4
 
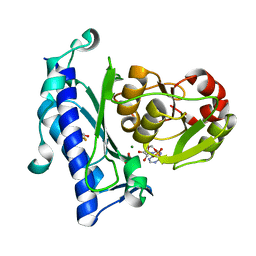 | | Crystal Structure of an Enzyme Displaying both Inositol-Polyphosphate 1-Phosphatase and 3'-Phosphoadenosine-5'-Phosphate Phosphatase Activities | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Patel, S, Yenush, L, Rodriguez, P.L, Serrano, R, Blundell, T.L. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of an enzyme displaying both inositol-polyphosphate-1-phosphatase and 3'-phosphoadenosine-5'-phosphate phosphatase activities: a novel target of lithium therapy.
J.Mol.Biol., 315, 2002
|
|
1K9Z
 
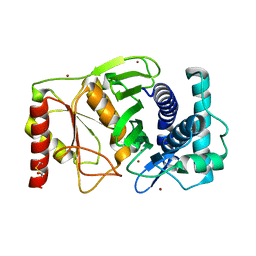 | |
1K9Y
 
 | | The PAPase Hal2p complexed with magnesium ions and reaction products: AMP and inorganic phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, Halotolerance protein HAL2, ... | | Authors: | Patel, S, Albert, A, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural enzymology of Li(+)-sensitive/Mg(2+)-dependent phosphatases.
J.Mol.Biol., 320, 2002
|
|
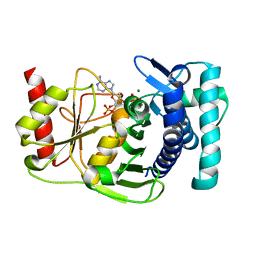
1KA1
 
 | | The PAPase Hal2p complexed with calcium and magnesium ions and reaction substrate: PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Patel, S, Albert, A, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural enzymology of Li(+)-sensitive/Mg(2+)-dependent phosphatases.
J.Mol.Biol., 320, 2002
|
|
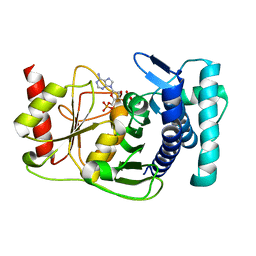
1KA0
 
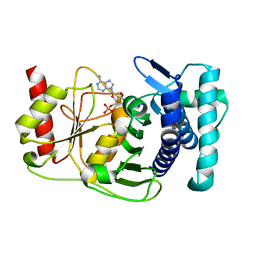 | |
1W50
 
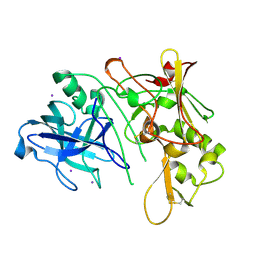 | | Apo Structure of BACE (Beta Secretase) | | Descriptor: | BETA-SECRETASE 1, IODIDE ION | | Authors: | Patel, S, Vuillard, L, Cleasby, A, Murray, C.W, Yon, J. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Apo and Inhibitor Complex Structures of Bace (Beta-Secretase)
J.Mol.Biol., 343, 2004
|
|
1W51
 
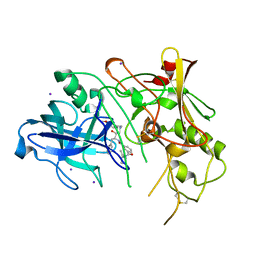 | | BACE (Beta Secretase) in complex with a nanomolar non-peptidic inhibitor | | Descriptor: | 3-[({(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}AMINO)(HYDROXY)METHYL]-N,N-DIPROPYLBENZAMIDE, BETA-SECRETASE 1, IODIDE ION | | Authors: | Patel, S, Vuillard, L, Cleasby, A, Murray, C.W, Yon, J. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Apo and Inhibitor Complex Structures of Bace (Beta-Secretase)
J.Mol.Biol., 343, 2004
|
|
5O57
 
 | | Solution Structure of the N-terminal Region of Dkk4 | | Descriptor: | Dickkopf-related protein 4 | | Authors: | Waters, L.C, Patel, S, Barkell, A.M, Muskett, F.W, Robinson, M.K, Holdsworth, G, Carr, M.D. | | Deposit date: | 2017-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of Dickkopf 4 (Dkk4): New insights into Dkk evolution and regulation of Wnt signaling by Dkk and Kremen proteins.
J. Biol. Chem., 293, 2018
|
|
2OHU
 
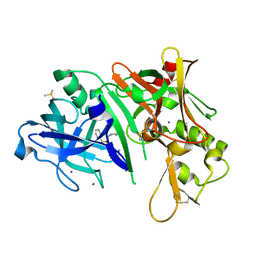 | | X-ray crystal structure of beta secretase complexed with compound 8b | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHR
 
 | | X-ray crystal structure of beta secretase complexed with compound 6a | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHP
 
 | | X-ray crystal structure of beta secretase complexed with compound 3 | | Descriptor: | 6-[2-(1H-INDOL-6-YL)ETHYL]PYRIDIN-2-AMINE, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHS
 
 | | X-ray crystal structure of beta secretase complexed with compound 6b | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHT
 
 | | X-ray crystal structure of beta secretase complexed with compound 7 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHQ
 
 | | X-ray crystal structure of beta secretase complexed with compound 4 | | Descriptor: | 6-[2-(3'-METHOXYBIPHENYL-3-YL)ETHYL]PYRIDIN-2-AMINE, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHL
 
 | |
2OHK
 
 | |
2OF0
 
 | | X-ray crystal structure of beta secretase complexed with compound 5 | | Descriptor: | (2S)-1-(2,5-dimethylphenoxy)-3-morpholin-4-ylpropan-2-ol, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-02 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to beta-Secretase.
J.Med.Chem., 50, 2007
|
|
2OHN
 
 | |
2OHM
 
 | |
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URL
 
 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URN
 
 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | Descriptor: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
1I57
 
 | | CRYSTAL STRUCTURE OF APO HUMAN PTP1B (C215S) MUTANT | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHO-TYROSINE PHOSPHATASE 1B | | Authors: | Scapin, G, Patel, S, Patel, V, Kennedy, B, Asante-Appiah, E. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of apo protein-tyrosine phosphatase 1B C215S mutant: more than just an S --> O change.
Protein Sci., 10, 2001
|
|
1LQF
 
 | | Structure of PTP1b in Complex with a Peptidic Bisphosphonate Inhibitor | | Descriptor: | N-BENZOYL-L-GLUTAMYL-[4-PHOSPHONO(DIFLUOROMETHYL)]-L-PHENYLALANINE-[4-PHOSPHONO(DIFLUORO-METHYL)]-L-PHENYLALANINEAMIDE, protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Asante-Appiah, E, Patel, S, Dufresne, C, Scapin, G. | | Deposit date: | 2002-05-10 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of PTP-1B in complex with a peptide inhibitor reveals an alternative binding mode for bisphosphonates.
Biochemistry, 41, 2002
|
|
4URO
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Novobiocin | | Descriptor: | DNA GYRASE SUBUNIT B, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
