2GP0
 
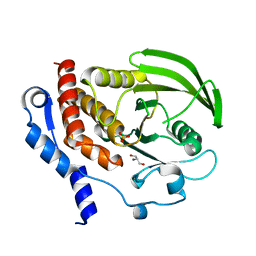 | | HePTP Catalytic Domain (residues 44-339), S225D mutant | | Descriptor: | GLYCEROL, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 7 | | Authors: | Page, R. | | Deposit date: | 2006-04-15 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | HePTP Catalytic Domain (residues 44-339), S225D mutant
To be Published
|
|
5SWF
 
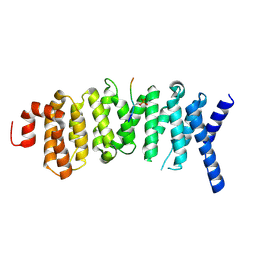 | | The structure of the PP2A B56 subunit double phosphorylated BubR1 complex | | Descriptor: | Double phosphorylated BubR1, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Page, R, Wang, X, Bajaj, R, Peti, W. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Expanding the PP2A Interactome by Defining a B56-Specific SLiM.
Structure, 24, 2016
|
|
5SW9
 
 | | The structure of the PP2A B56 subunit RepoMan complex | | Descriptor: | RepoMan, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Page, R, Wang, X, Bajaj, R, Peti, W. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.846 Å) | | Cite: | Expanding the PP2A Interactome by Defining a B56-Specific SLiM.
Structure, 24, 2016
|
|
5W1E
 
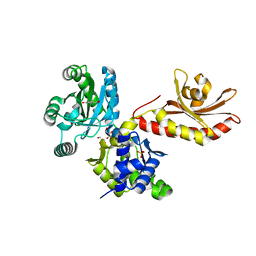 | | PobR in complex with PHB | | Descriptor: | GLYCEROL, P-HYDROXYBENZOIC ACID, Putative transcriptional regulator, ... | | Authors: | Page, R, Peti, W, Lord, D.M, Bajaj, R, Zhang, R. | | Deposit date: | 2017-06-02 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A peculiar IclR family transcription factor regulates para-hydroxybenzoate catabolism in Streptomyces coelicolor.
Nucleic Acids Res., 46, 2018
|
|
3V4Y
 
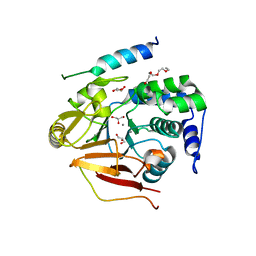 | | Crystal Structure of the first Nuclear PP1 holoenzyme | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Nuclear inhibitor of protein phosphatase 1, ... | | Authors: | Page, R, Peti, W, O'Connell, N.E, Nichols, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
Structure, 20, 2012
|
|
4PCV
 
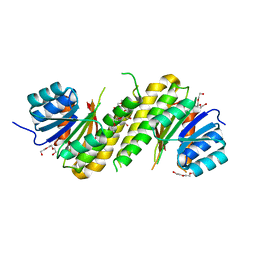 | | The structure of BdcA (YjgI) from E. coli | | Descriptor: | BdcA (YjgI), PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Page, R, Peti, W, Lord, D. | | Deposit date: | 2014-04-16 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BdcA, a protein important for Escherichia coli biofilm dispersal, is a short-chain dehydrogenase/reductase that binds specifically to NADPH.
Plos One, 9, 2014
|
|
6ATD
 
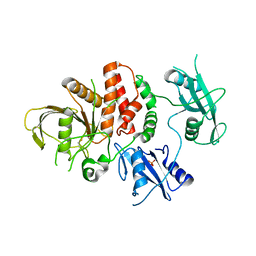 | |
2HVL
 
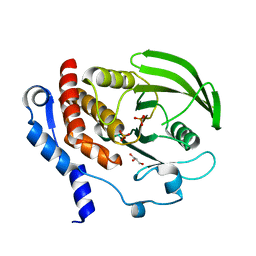 | |
1ZC0
 
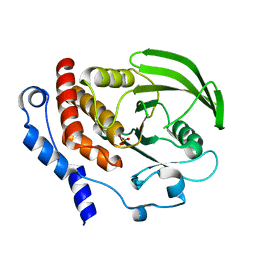 | |
2OXL
 
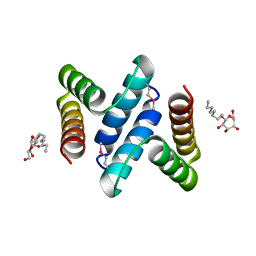 | | Structure and Function of the E. coli Protein YmgB: a Protein Critical for Biofilm Formation and Acid Resistance | | Descriptor: | Hypothetical protein ymgB, octyl beta-D-glucopyranoside | | Authors: | Page, R, Peti, W, Woods, T.K, Palermino, J.M, Doshi, O. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid-resistance.
J.Mol.Biol., 373, 2007
|
|
3FMY
 
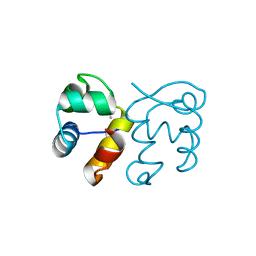 | |
5K6S
 
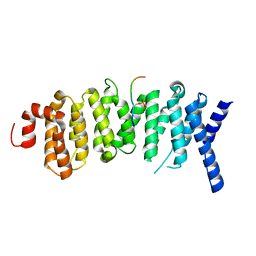 | | The structure of the PP2A B56 subunit BubR1 complex | | Descriptor: | BubR1, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Page, R, Wang, X, Bajaj, R, Peti, W. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Expanding the PP2A Interactome by Defining a B56-Specific SLiM.
Structure, 24, 2016
|
|
2QDM
 
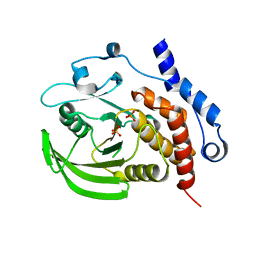 | |
8U5G
 
 | | Crystal structure of the co-expressed SDS22:PP1:I3 complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-09-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The SDS22:PP1:I3 complex: SDS22 binding to PP1 loosens the active site metal to prime metal exchange.
J.Biol.Chem., 300, 2023
|
|
4MOV
 
 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4503 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5J28
 
 | | Ki67-PP1g (protein phosphatase 1, gamma isoform) holoenzyme complex | | Descriptor: | Antigen KI-67, MALONATE ION, SODIUM ION, ... | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
4MOY
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP0
 
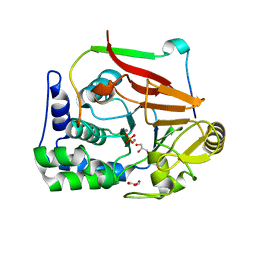 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4XPN
 
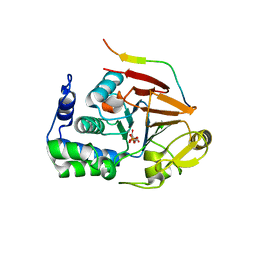 | | Crystal Structure of Protein Phosphate 1 complexed with PP1 binding domain of GADD34 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Structural and Functional Analysis of the GADD34:PP1 eIF2 alpha Phosphatase.
Cell Rep, 11, 2015
|
|
4Y14
 
 | | Structure of protein tyrosine phosphatase 1B complexed with inhibitor (PTP1B:CPT157633) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-bromo-4-[difluoro(phosphono)methyl]-N-methyl-Nalpha-(methylsulfonyl)-L-phenylalaninamide, CHLORIDE ION, ... | | Authors: | Choy, M.S, Connors, C, Page, R, Peti, W. | | Deposit date: | 2015-02-06 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PTP1B inhibition suggests a therapeutic strategy for Rett syndrome.
J.Clin.Invest., 125, 2015
|
|
7T0Y
 
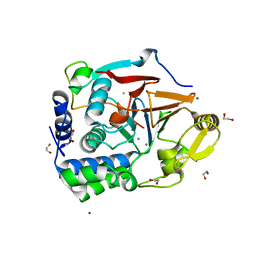 | | The Ribosomal RNA Processing 1B Protein Phosphatase-1 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, FLUORIDE ION, ... | | Authors: | Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The ribosomal RNA processing 1B:protein phosphatase 1 holoenzyme reveals non-canonical PP1 interaction motifs.
Cell Rep, 41, 2022
|
|
4PHL
 
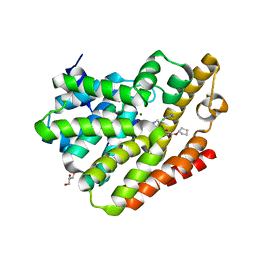 | | TbrPDEB1-inhibitor complex | | Descriptor: | 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, Class 1 phosphodiesterase PDEB1, ETHANOL, ... | | Authors: | Choy, M.S, Bland, N, Peti, W, Page, R. | | Deposit date: | 2014-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TbrPDEB1-inhibitor complex
To Be Published
|
|
5SVE
 
 | | Structure of Calcineurin in complex with NFATc1 LxVP peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Sheftic, S.R, Page, R, Peti, W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Investigating the human Calcineurin Interaction Network using the pi LxVP SLiM.
Sci Rep, 6, 2016
|
|
4P9F
 
 | | E. coli McbR/YncC | | Descriptor: | HTH-type transcriptional regulator mcbR | | Authors: | Lord, D.M, Page, R, Peti, W. | | Deposit date: | 2014-04-03 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | McbR/YncC: Implications for the Mechanism of Ligand and DNA Binding by a Bacterial GntR Transcriptional Regulator Involved in Biofilm Formation.
Biochemistry, 53, 2014
|
|
8SO0
 
 | | Cryo-EM structure of the PP2A:B55-FAM122A complex | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-04-28 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79961 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
