3JTN
 
 | | Crystal Structure of the c-terminal domain of YpbH | | Descriptor: | Adapter protein mecA 2, IODIDE ION | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the MecA degradation tag
To be Published
|
|
3JTP
 
 | | crystal structure of the C-terminal domain of MecA | | Descriptor: | Adapter protein mecA 1, IODIDE ION | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | crystal structure of the MecA degradation tag
To be Published
|
|
3JTO
 
 | | Crystal structure of the c-terminal domain of YpbH | | Descriptor: | Adapter protein mecA 2 | | Authors: | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the MecA Degradation Tag
To be Published
|
|
3J3T
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
3J3S
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J3U
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
2Y1R
 
 | | Structure of MecA121 & ClpC N-domain complex | | Descriptor: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
2Y1Q
 
 | | Crystal Structure of ClpC N-terminal Domain | | Descriptor: | NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, SULFATE ION | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
3PXG
 
 | | Structure of MecA121 and ClpC1-485 complex | | Descriptor: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.654 Å) | | Cite: | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
7BV3
 
 | | Crystal structure of a ugt transferase from Siraitia grosvenorii in complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases
Green Synth Catal, 2021
|
|
6IIK
 
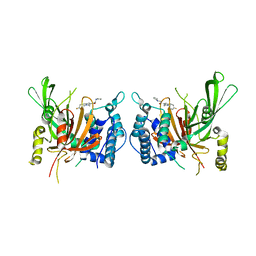 | | USP14 catalytic domain with IU1 | | Descriptor: | 1-[1-(4-fluorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(pyrrolidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, He, W, Wang, F. | | Deposit date: | 2018-10-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6IIN
 
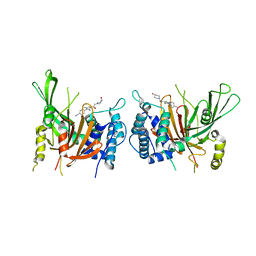 | | USP14 catalytic domain with IU1-248 | | Descriptor: | 4-{3-[(4-hydroxypiperidin-1-yl)acetyl]-2,5-dimethyl-1H-pyrrol-1-yl}benzonitrile, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6IIL
 
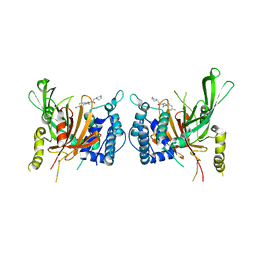 | | USP14 catalytic domain bind to IU1-47 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(piperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, Wang, F, Wang, J.W, He, W, Ding, S, Li, J.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6IIM
 
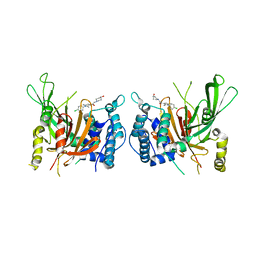 | | USP14 catalytic domain with IU1-206 | | Descriptor: | 1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(4-hydroxypiperidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, J.W, Wang, F, Wang, Y.W. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
6K9N
 
 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6KBE
 
 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
3JCO
 
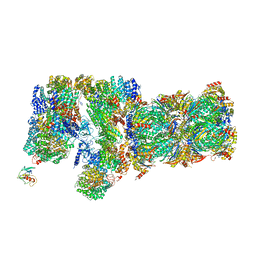 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8HOG
 
 | | Crystal structure of Bcl-2 in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOI
 
 | | Crystal structure of Bcl-2 D103Y in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, FORMIC ACID, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
6ISU
 
 | | Crystal structure of Lys27-linked di-ubiquitin in complex with its selective interacting protein UCHL3 | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Ding, S, Pan, M, Zheng, Q, Ren, Y, Hong, D. | | Deposit date: | 2018-11-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
