6X0O
 
 | | Single-Particle Cryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB | | Authors: | Tan, Y.Z, Rodrigues, J, Keener, J.E, Zheng, R.B, Brunton, R, Kloss, B, Giacometti, S.I, Rosario, A.L, Zhang, L, Niederweis, M, Clarke, O.B, Lowary, T.L, Marty, M.T, Archer, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis.
Nat Commun, 11, 2020
|
|
7MJS
 
 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
5DS1
 
 | | Core domain of the class II small heat-shock protein HSP 17.7 from Pisum sativum | | Descriptor: | 17.1 kDa class II heat shock protein | | Authors: | Hochberg, G.K.A, Laganoswky, A, Allison, T.A, Shepherd, D.A, Benesch, J.L.P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions.
Science, 359, 2018
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
6WTT
 
 | | Crystals Structure of the SARS-CoV-2 (COVID-19) main protease with inhibitor GC-376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Sacco, M, Ma, C, Chen, Y, Wang, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease.
Cell Res., 30, 2020
|
|
5DS2
 
 | | Core domain of the class I small heat-shock protein HSP 18.1 from Pisum sativum | | Descriptor: | 18.1 kDa class I heat shock protein, SULFATE ION | | Authors: | Shepherd, D.A, Laganowsky, A, Allison, T.M, Hochberg, G.K.A, Benesch, J.L.P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions.
Science, 359, 2018
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
6XFN
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW243 | | Descriptor: | 3C-like proteinase, GLYCEROL, UAW243 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-15 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
7RN0
 
 | |
7RN1
 
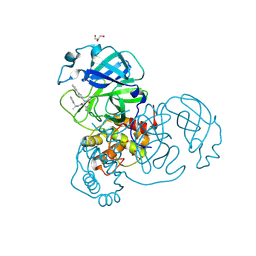 | |
7RCZ
 
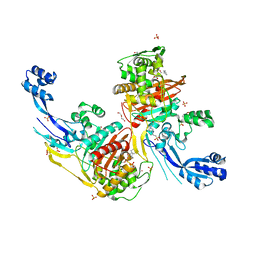 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RD0
 
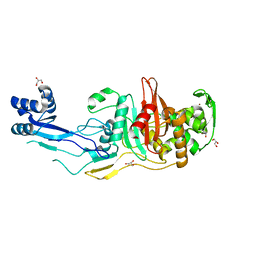 | |
7RCY
 
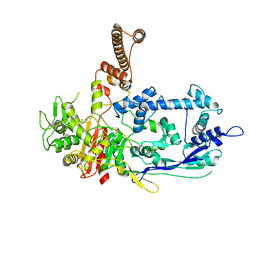 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCX
 
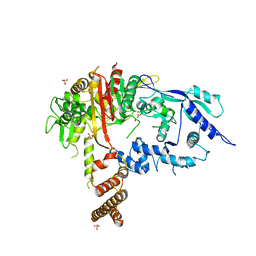 | |
7RCW
 
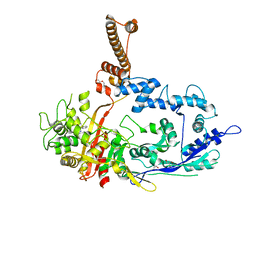 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
