5NFR
 
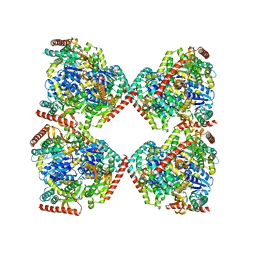 | | Crystal structure of malate dehydrogenase from Plasmodium falciparum (PfMDH) | | Descriptor: | CITRIC ACID, Malate dehydrogenase | | Authors: | Lunev, S, Romero, A.R, Batista, F.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-03-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oligomeric interfaces as a tool in drug discovery: Specific interference with activity of malate dehydrogenase of Plasmodium falciparum in vitro.
PLoS ONE, 13, 2018
|
|
6FBA
 
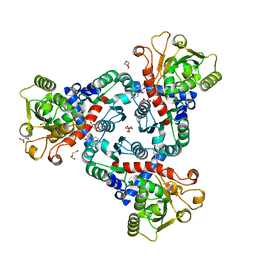 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with bound inhibitor 2,3-naphthalenediol | | Descriptor: | Aspartate transcarbamoylase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.A, Wang, C, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a non-competitive inhibitor of Plasmodium falciparum aspartate transcarbamoylase.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5ILQ
 
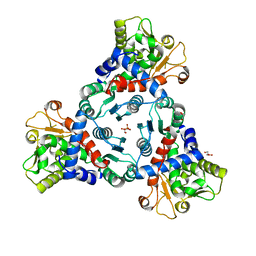 | | Crystal structure of truncated unliganded Aspartate Transcarbamoylase from Plasmodium falciparum | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, SULFATE ION | | Authors: | Lunev, S, Bosch, S.S, Batista, F.D.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2016-03-04 | | Release date: | 2016-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of truncated aspartate transcarbamoylase from Plasmodium falciparum.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ILN
 
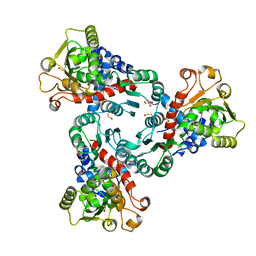 | | Crystal structure of Aspartate Transcarbamoylase from Plasmodium falciparum (PfATC) with bound citrate | | Descriptor: | Aspartate carbamoyltransferase, CITRATE ANION, GLYCEROL, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.D.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of truncated Aspartate Transcarbamoylase from Plasmodium falciparum
To be published
|
|
5MC7
 
 | |
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
6R8G
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with 4-(3,4-difluorophenyl)thiazol-2-amine | | Descriptor: | 4-[3,4-bis(fluoranyl)phenyl]-1,3-thiazol-2-amine, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fragment-Based Approach Identifies an Allosteric Pocket that Impacts Malate Dehydrogenase Activity
Commun Biol, 2021
|
|
6HL7
 
 | | Crystal structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with mutated active site (R109A/K138A) and N-carbamoyl-L-phosphate bound | | Descriptor: | Aspartate transcarbamoylase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Bosch, S.S, Lunev, S, Wrenger, C, Groves, M.R. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Target Validation of Aspartate Transcarbamoylase fromPlasmodium falciparumby Torin 2.
Acs Infect Dis., 6, 2020
|
|
5NKZ
 
 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
6ZRT
 
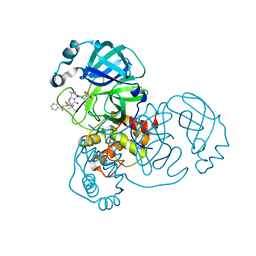 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, DIMETHYL SULFOXIDE, Main Protease | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
5NL8
 
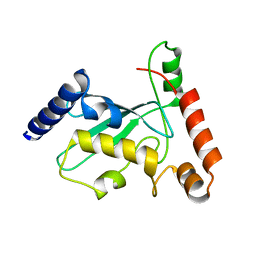 | | Pex4 of Hansenula Polymorpha | | Descriptor: | Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Groves, M, Williams, C. | | Deposit date: | 2017-04-04 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
6ZRU
 
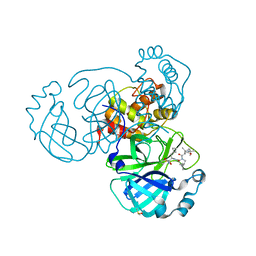 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Boceprevir | | Descriptor: | DIMETHYL SULFOXIDE, Main Protease, boceprevir (bound form) | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
