7YRB
 
 | | UBR box of human UBR6 | | Descriptor: | F-box protein 11, isoform CRA_f, SULFATE ION, ... | | Authors: | Kim, B, Song, H.K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of UBR box from human UBR6
To Be Published
|
|
9ISP
 
 | | DUF2436 domain which is frequently found in virulence proteins from Porphyromonas gingivalis | | Descriptor: | Protease PrtH | | Authors: | Kim, B, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2024-07-18 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | First crystal structure of the DUF2436 domain of virulence proteins from Porphyromonas gingivalis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
7C1C
 
 | |
7C1A
 
 | |
7C18
 
 | |
7CVX
 
 | |
7CVW
 
 | |
7CVV
 
 | |
7CVU
 
 | |
1PVE
 
 | |
4GRG
 
 | |
2ABY
 
 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
6B8C
 
 | |
8IS3
 
 | | Structural model for the micelle-bound indolicidin-like peptide in solution | | Descriptor: | Indolicidin-like antimicrobial peptide | | Authors: | Kim, B, Ko, Y.H, Kim, J, Lee, J, Nam, C.H, Kim, J.H. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural model for the micelle-bound indolicidin-like peptide in solution
To Be Published
|
|
6A9P
 
 | |
7D7O
 
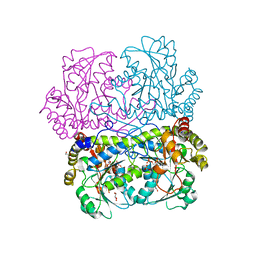 | | Crystal structure of cystathionine gamma-lyase from Bacillus cereus ATCC 14579 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sagong, H.-Y, Kim, B, Kim, K.-J. | | Deposit date: | 2020-10-05 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and Functional Characterization of Cystathionine gamma-lyase from Bacillus cereus ATCC 14579.
J.Agric.Food Chem., 68, 2020
|
|
6PWY
 
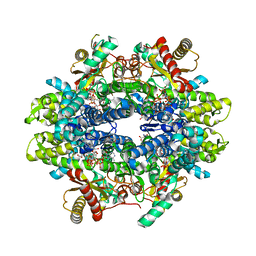 | | Structure of C. elegans ZK177.8, SAMHD1 ortholog | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lim, C.S, Maehigashi, T, Wade, L.R, Bowen, N, Knecht, K, Xiong, Y, Kim, B. | | Deposit date: | 2019-07-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | ZK177.8 of Caenorhabditis elegans:
Aicardi-Goutieres Syndrome SAMHD1 Ortholog
To Be Published
|
|
2ZNF
 
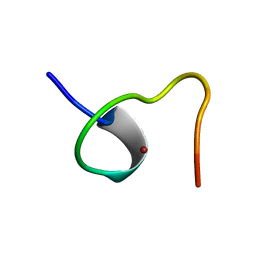 | | HIGH-RESOLUTION STRUCTURE OF AN HIV ZINC FINGERLIKE DOMAIN VIA A NEW NMR-BASED DISTANCE GEOMETRY APPROACH | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Summers, M.F, South, T.L, Kim, B, Hare, D.R. | | Deposit date: | 1990-03-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach.
Biochemistry, 29, 1990
|
|
6ETJ
 
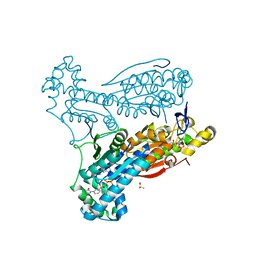 | | HUMAN PFKFB3 IN COMPLEX WITH KAN0438241 | | Descriptor: | 4-[[3-(5-fluoranyl-2-oxidanyl-phenyl)phenyl]sulfonylamino]-2-oxidanyl-benzoic acid, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Gustafsson, N.M.S, Lundback, T, Farnegardh, K, Groth, P, Wiitta, E, Jonsson, M, Hallberg, K, Pennisi, R, Huguet Ninou, A, Martinsson, J, Norstrom, C, Schultz, J, Andersson, M, Markova, N, Marttila, P, Norin, M, Olin, T, Helleday, T. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination.
Nat Commun, 9, 2018
|
|
8A1Q
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor STP0404 (Pirmitegravir) | | Descriptor: | (2S)-tert-butoxy{4-(4-chlorophenyl)-2,3,6-trimethyl-1-[(1-methyl-1H-pyrazol-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridin-5-yl}acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
8A1P
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BI-D | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
7KE0
 
 | |
8S9Q
 
 | |
8T5B
 
 | |
8T5A
 
 | |
