4NIM
 
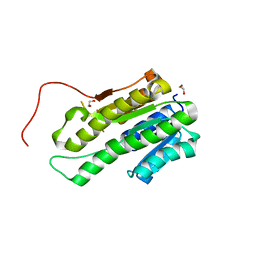 | |
4NPC
 
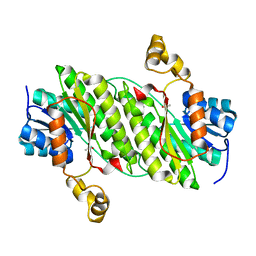 | | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis | | Descriptor: | ACETATE ION, Sorbitol dehydrogenase | | Authors: | Dranow, D.M, Davies, D.R, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis
To be Published
|
|
4NI5
 
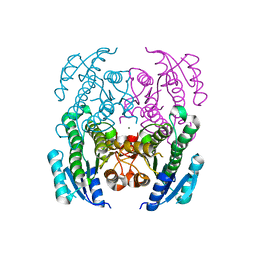 | | Crystal Structure of a Short Chain Dehydrogenase from Brucella suis | | Descriptor: | MAGNESIUM ION, Oxidoreductase, short-chain dehydrogenase/reductase family protein | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Short Chain Dehydrogenase from Brucella suis
TO BE PUBLISHED
|
|
4NOZ
 
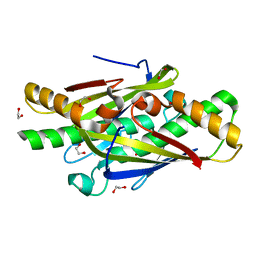 | | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Organic hydroperoxide resistance protein | | Authors: | Dranow, D.M, Lukacs, C.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia
TO BE PUBLISHED
|
|
4QTP
 
 | | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Anti-sigma factor antagonist, CITRIC ACID, ... | | Authors: | Dranow, D.M, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of an Anti-sigma Factor Antagonist from Mycobacterium paratuberculosis
TO BE PUBLISHED
|
|
6CHD
 
 | |
6Q0D
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00384414-01 AT 2.05 A RESOLUTION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-[3-(cyclopentylethynyl)-4-fluorophenyl]-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Dranow, D.M, Davies, D.R. | | Deposit date: | 2019-08-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
5IFU
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with Glyburide | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, CHLORIDE ION, ... | | Authors: | Dranow, D.M, Hewitt, S.N, Abendroth, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and Structural Characterization of Selective Allosteric Inhibitors of the Plasmodium falciparum Drug Target, Prolyl-tRNA-synthetase.
ACS Infect Dis, 3, 2017
|
|
4NPS
 
 | | Crystal Structure of Bep1 protein (VirB-translocated Bartonella effector protein) from Bartonella clarridgeiae | | Descriptor: | ACETATE ION, Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolutionary Diversification of Host-Targeted Bartonella Effectors Proteins Derived from a Conserved FicTA Toxin-Antitoxin Module.
Microorganisms, 9, 2021
|
|
5W8L
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 59 and NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-([1,1'-biphenyl]-3-yl)-5-(cyclopropylmethyl)-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8H
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 11 | | Descriptor: | 2-[3-(4-fluorophenyl)-5-(trifluoromethyl)-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
4TMD
 
 | | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis | | Descriptor: | IODIDE ION, Uncharacterized protein | | Authors: | Horanyi, P.S, Dranow, D.M, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-06-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis
To Be Published
|
|
6Q13
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00420737-09 AT 2.00 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-[5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-3-{3-[(5-methylthiophen-2-yl)ethynyl]phenyl}-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2019-08-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
4WJB
 
 | | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Putative amidohydrolase/peptidase, SULFATE ION, ... | | Authors: | Lukacs, C.M, Dranow, D.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia
To Be Published
|
|
4WMW
 
 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMS
 
 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMX
 
 | | The structure of MBP-MCL1 bound to ligand 6 at 2.0A | | Descriptor: | 4-ethenyl-2-[(phenylsulfonyl)amino]benzoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMT
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 1 AT 2.35A | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
5W8K
 
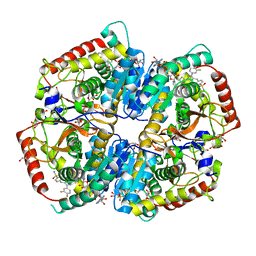 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
6DAV
 
 | | Crystal Structure of Human DHFR complexed with NADP and N10formyltetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase, N-(4-{[(2,4-diaminopteridin-6-yl)methyl](hydroxymethyl)amino}benzene-1-carbonyl)-L-glutamic acid, ... | | Authors: | Mayclin, S.J, Dranow, D.M, Lorimer, D.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human DHFR complexed with NADP and N10formyltetrahydrofolate
to be published
|
|
5V8T
 
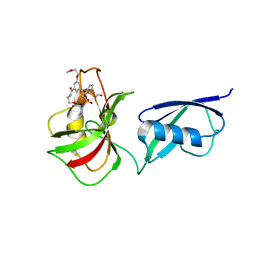 | | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354 | | Descriptor: | 2-{[3,5-bis(2-methoxyethoxy)benzene-1-carbonyl]amino}ethyl (2S)-1-(benzylsulfonyl)piperidine-2-carboxylate, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Lorimer, D.D, Dranow, D.M, Seufert, F, Abendroth, J, Holzgrabe, U. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354
to be published
|
|
5TQJ
 
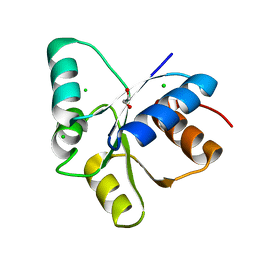 | |
8DUY
 
 | |
4MAQ
 
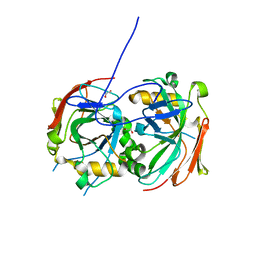 | |
