6F27
 
 | | NMR solution structure of non-bound [des-Arg10]-kallidin (DAKD) | | Descriptor: | DAKD | | Authors: | Richter, C, Jonker, H.R.A, Schwalbe, H, Joedicke, L, Mao, J, Kuenze, G, Reinhart, C, Kalavacherla, T, Meiler, J, Preu, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3Y
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide bound to human Bradykinin 1 Receptor (B1R) determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3X
 
 | | Backbone structure of Des-Arg10-Kallidin (DAKD) peptide in frozen DDM/CHS detergent micelle solution determined by DNP-enhanced MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Kuenze, G, Joedicke, L, Meiler, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
2FHG
 
 | | Crystal Structure of Mycobacterial Tuberculosis Proteasome | | Descriptor: | 20S proteasome, alpha and beta subunits, proteasome, ... | | Authors: | Hu, G, Lin, G, Wang, M, Dick, L, Xu, R.M, Nathan, C, Li, H. | | Deposit date: | 2005-12-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate.
Mol.Microbiol., 59, 2006
|
|
6F3V
 
 | | Backbone structure of bradykinin (BK) peptide bound to human Bradykinin 2 Receptor (B2R) determined by MAS SSNMR | | Descriptor: | Bradykinin (BK) | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3W
 
 | | Backbone structure of free bradykinin (BK) in DDM/CHS detergent micelle determined by MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
2FHH
 
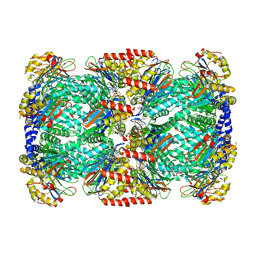 | |
5TR4
 
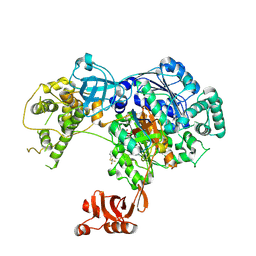 | | Structure of Ubiquitin activating enzyme (Uba1) in complex with ubiquitin and TAK-243 | | Descriptor: | Ubiquitin, Ubiquitin-activating enzyme E1 1, [(1~{R},2~{R},3~{S},4~{R})-2,3-bis(oxidanyl)-4-[[2-[3-(trifluoromethylsulfanyl)phenyl]pyrazolo[1,5-a]pyrimidin-7-yl]amino]cyclopentyl]methyl sulfamate | | Authors: | Sintchak, M.D. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and Characterization of a Small Molecule Inhibitor of the Ubiquitin Activating Enzyme (TAK-243)
To be published
|
|
1R4L
 
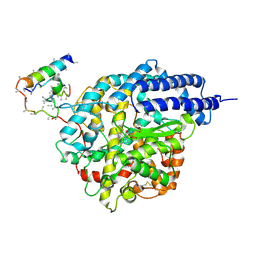 | | Inhibitor Bound Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | (S,S)-2-{1-CARBOXY-2-[3-(3,5-DICHLORO-BENZYL)-3H-IMIDAZOL-4-YL]-ETHYLAMINO}-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1R42
 
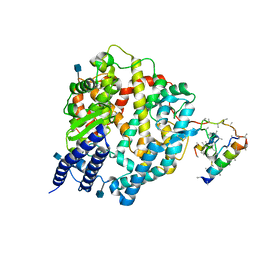 | | Native Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ZINC ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
3M93
 
 | |
6GSY
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3 | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
6GSV
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
1CKP
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR PURVALANOL B | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN (CYCLIN-DEPENDENT PROTEIN KINASE 2), PURVALANOL B | | Authors: | Gray, N.S, Thunnissen, A.M.W.H, Schultz, P.G, Kim, S.H. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors.
Science, 281, 1998
|
|
1DM2
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR HYMENIALDISINE | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Thunnissen, A.M, Kim, S.-H. | | Deposit date: | 1999-12-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of cyclin-dependent kinases, GSK-3beta and CK1 by hymenialdisine, a marine sponge constituent.
Chem.Biol., 7, 2000
|
|
2GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
3M94
 
 | | Complex crystal structure of Ascaris suum eIF4E-3 with m2,2,7G cap | | Descriptor: | ACETYL GROUP, Eukaryotic translation initiation factor 4E-binding protein 1, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Liu, W, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2010-03-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nematode eIF4E binding an m2,2,7G-Cap and its implications for translation initiation.
Nucleic Acids Res., 39, 2011
|
|
3GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
1PGT
 
 | |
1GSQ
 
 | | THREE-DIMENSIONAL STRUCTURE, CATALYTIC PROPERTIES AND EVOLUTION OF A SIGMA CLASS GLUTATHIONE TRANSFERASE FROM SQUID, A PROGENITOR OF THE LENS-CRYSTALLINS OF CEPHALOPODS | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Ji, X, Rosenvinge, E.C.V, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1995-01-09 | | Release date: | 1995-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure, catalytic properties, and evolution of a sigma class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods.
Biochemistry, 34, 1995
|
|
1GSU
 
 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | Descriptor: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | Deposit date: | 1997-09-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
6GSU
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
6GSW
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
6GSX
 
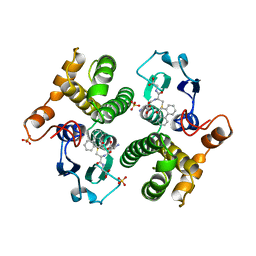 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
6GST
 
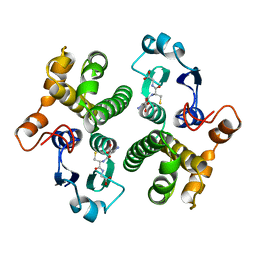 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3 | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
