3V3R
 
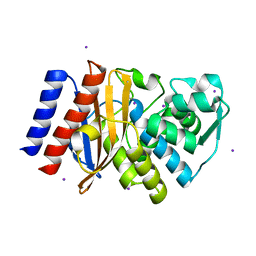 | | Crystal Structure of GES-11 | | Descriptor: | Extended spectrum class A beta-lactamase GES-11, IODIDE ION, SODIUM ION | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Kinetic and crystallographic studies of extended-spectrum GES-11, GES-12, and GES-14 beta-lactamases.
Antimicrob.Agents Chemother., 56, 2012
|
|
1IGQ
 
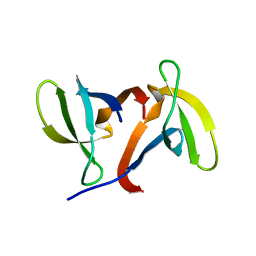 | |
1IGU
 
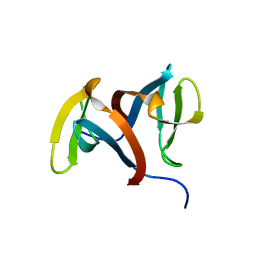 | |
3SW3
 
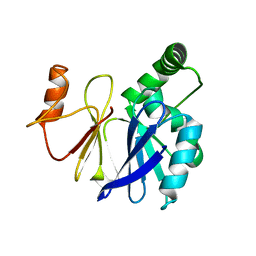 | |
3TSG
 
 | | Crystal structure of GES-14 | | Descriptor: | Extended-spectrum beta-lactamase GES-14, GLYCEROL, IODIDE ION | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and crystallographic studies of extended-spectrum GES-11, GES-12, and GES-14 beta-lactamases.
Antimicrob.Agents Chemother., 56, 2012
|
|
3T9M
 
 | |
3V3S
 
 | | Crystal structure of GES-18 | | Descriptor: | Extended spectrum beta-lactamase GES-18 | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GES-18, a new carbapenem-hydrolyzing GES-Type beta-lactamase from pseudomonas aeruginosa that contains Ile80 and Ser170 residues.
Antimicrob.Agents Chemother., 57, 2013
|
|
3IOG
 
 | | Crystal structure of CphA N220G mutant with inhibitor 18 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION, ... | | Authors: | Delbruck, H, Bebrone, C, Hoffmann, K.M.V. | | Deposit date: | 2009-08-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-beta-lactamases
J.Med.Chem., 53, 2010
|
|
3IOF
 
 | | Crystal structure of CphA N220G mutant with inhibitor 10a | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION, ... | | Authors: | Delbruck, H, Bebrone, C, Hoffmann, K.M.V. | | Deposit date: | 2009-08-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-beta-lactamases
J.Med.Chem., 53, 2010
|
|
3F9O
 
 | |
3FAI
 
 | | The Di Zinc Carbapenemase CphA N220G mutant | | Descriptor: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Delbruck, H, Hoffmann, K.M.V. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of the dizinc subclass B2 metallo-beta-lactamase CphA reveals that the second inhibitory zinc ion binds in the histidine site
Antimicrob.Agents Chemother., 53, 2009
|
|
2WRS
 
 | | Crystal Structure of the Mono-Zinc Metallo-beta-lactamase VIM-4 from Pseudomonas aeruginosa | | Descriptor: | BETA-LACTAMASE VIM-4, CITRATE ANION, CITRIC ACID, ... | | Authors: | Lassaux, P, Hamel, M, Gulea, M, Delbruck, H, Traore, D.A.K, Mercuri, P.S, Horsfall, L, Dehareng, D, Gaumont, A.-C, Frere, J.-M, Ferrer, J.-L, Hoffmann, K, Galleni, M, Bebrone, C. | | Deposit date: | 2009-09-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
1I5F
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD-SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-02-27 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZ9
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZA
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZB
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZC
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1SAW
 
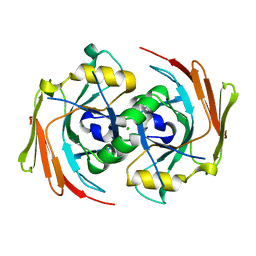 | | X-ray structure of homo sapiens protein FLJ36880 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, hypothetical protein FLJ36880 | | Authors: | Manjasetty, B.A, Niesen, F.H, Delbrueck, H, Goetz, F, Sievert, V, Buessow, K, Behlke, J, Heinemann, U. | | Deposit date: | 2004-02-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of fumarylacetoacetate hydrolase family member Homo sapiens FLJ36880.
Biol.Chem., 385, 2004
|
|
1ONI
 
 | | Crystal structure of a human p14.5, a translational inhibitor reveals different mode of ligand binding near the invariant residues of the Yjgf/UK114 protein family | | Descriptor: | 14.5 kDa translational inhibitor protein, BENZOIC ACID | | Authors: | Manjasetty, B.A, Delbrueck, H, Mueller, U, Erdmann, M.F, Heinemann, U. | | Deposit date: | 2003-02-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Homo sapiens protein hp14.5.
Proteins, 54, 2004
|
|
