5D0J
 
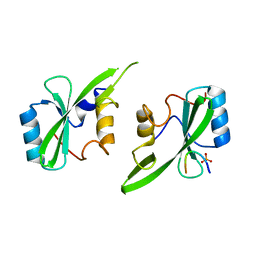 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
1MNT
 
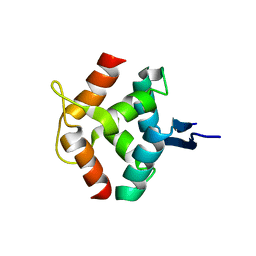 | | SOLUTION STRUCTURE OF DIMERIC MNT REPRESSOR (1-76) | | Descriptor: | MNT REPRESSOR | | Authors: | Burgering, M.J.M, Boelens, R, Gilbert, D.E, Breg, J.N, Knight, K.L, Sauer, R.T, Kaptein, R. | | Deposit date: | 1994-06-28 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dimeric Mnt repressor (1-76).
Biochemistry, 33, 1994
|
|
1ADZ
 
 | |
5EEL
 
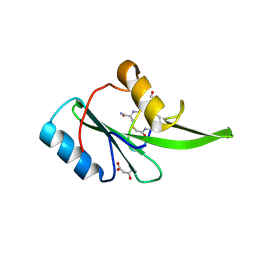 | | Grb7 SH2 with bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, FORMIC ACID, Growth factor receptor-bound protein 7, ... | | Authors: | Watson, G.M, Gunzburg, M.J, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
5VYE
 
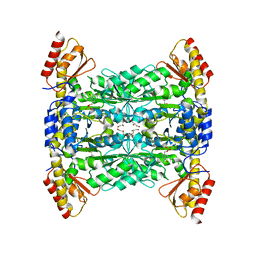 | |
5VG3
 
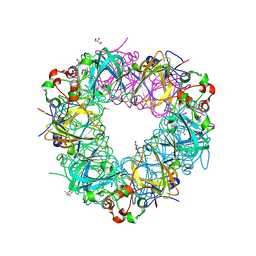 | |
6UFI
 
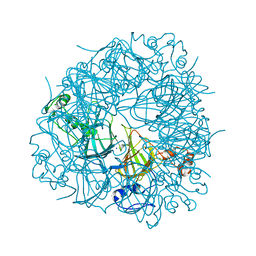 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
6TZP
 
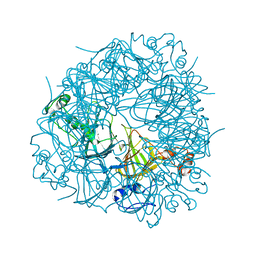 | | W96F Oxalate Decarboxylase (B. subtilis) | | Descriptor: | MANGANESE (II) ION, Oxalate decarboxylase | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-08-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
1ARR
 
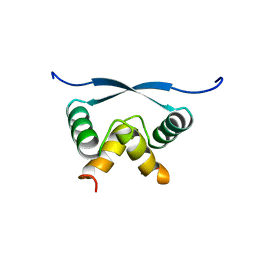 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
1ARQ
 
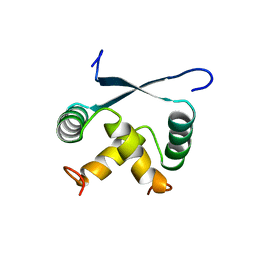 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
4J1Y
 
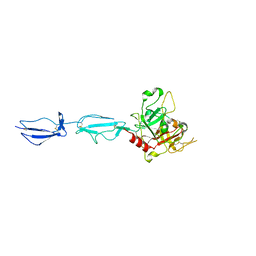 | | The X-ray crystal structure of human complement protease C1s zymogen | | Descriptor: | Complement C1s subcomponent | | Authors: | Perry, A.J, Wijeyewickrema, L.C, Wilmann, P.G, Gunzburg, M.J, D'Andrea, L, Irving, J.A, Pang, S.S, Duncan, R.C, Wilce, J.A, Whisstock, J.C, Pike, R.N. | | Deposit date: | 2013-02-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6645 Å) | | Cite: | A Molecular Switch Governs the Interaction between the Human Complement Protease C1s and Its Substrate, Complement C4.
J.Biol.Chem., 288, 2013
|
|
1JVI
 
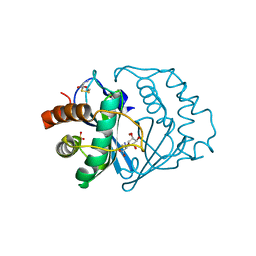 | | THE 2.2 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/RIBOSILHOMOCYSTEINE COMPLEX | | Descriptor: | (2S)-2-amino-4-[[(2S,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methylsulfanyl]butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ... | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1J98
 
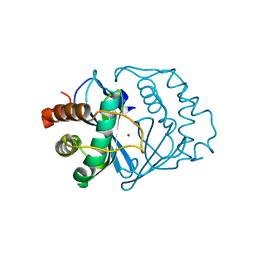 | | The 1.2 Angstrom Structure of Bacillus subtilis LuxS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A Structure of a Novel Quorum-Sensing Protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1JQW
 
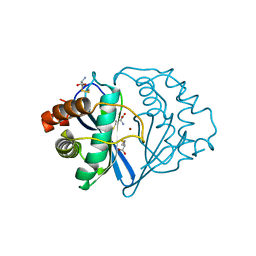 | | THE 2.3 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/HOMOCYSTEINE COMPLEX | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
4LT1
 
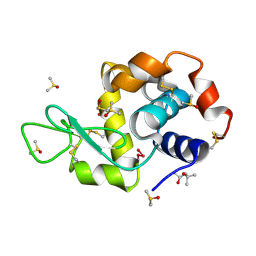 | | HEWL co-crystallised with Carboplatin in non-NaCl conditions: crystal 1 processed using the XDS software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboplatin binding to a model protein in non-NaCl conditions to eliminate partial conversion to cisplatin, and the use of different criteria to choose the resolution limit
To be Published
|
|
4LT2
 
 | | HEWL co-crystallized with Carboplatin in non-NaCl conditions: crystal 2 processed using the EVAL software package | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carboplatin binding to a model protein in non-NaCl conditions to eliminate partial conversion to cisplatin, and the use of different criteria to choose the resolution limit
To be Published
|
|
4B41
 
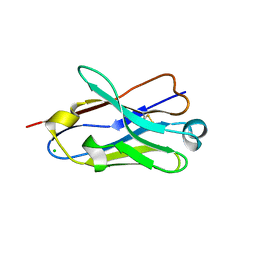 | | Crystal structure of an amyloid-beta binding single chain antibody G7 | | Descriptor: | ANTIBODY G7, CHLORIDE ION, GLYCEROL | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-07-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Crystal Structure of an Amyloid-Beta Binding Single Chain Antibody G7
To be Published
|
|
4BDX
 
 | |
4B5E
 
 | | Crystal Structure of an amyloid-beta binding single chain antibody PS2-8 | | Descriptor: | GLYCEROL, PS2-8, SULFATE ION | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Amyloid-Beta Binding Single Chain Antibody Ps2-8
To be Published
|
|
4BDW
 
 | |
1TFX
 
 | |
5EEQ
 
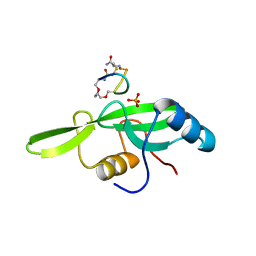 | | Grb7 SH2 with the G7-B1 bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Ambaye, N.D, Watson, G.M, Wilce, M.C.J, Wilce, G.M. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
7SO0
 
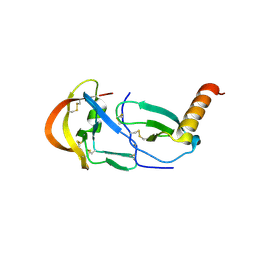 | | Crystal Structure of the Engineered Tick Evasin EVA-P974(F31A) Complexed to Human Chemokine CCL2 | | Descriptor: | C-C motif chemokine 2, Evasin P974 | | Authors: | Bhusal, R.P, Devkota, S.R, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-guided engineering of tick evasins for targeting chemokines in inflammatory diseases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KBG
 
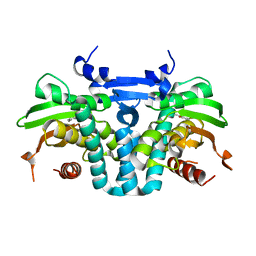 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH OCRESOL | | Descriptor: | MopR, ZINC ION, o-cresol | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBE
 
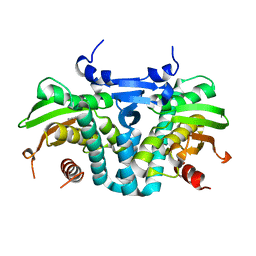 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH PHENOL | | Descriptor: | MopR, PHENOL, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
