8HUC
 
 | | Crystal structure of PaIch (Pec1) | | Descriptor: | GLYCEROL, MaoC_dehydrat_N domain-containing protein, NITRATE ION | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural and functional insights of itaconyl-CoA hydratase from Pseudomonas aeruginosa highlight a novel N-terminal hotdog fold.
Febs Lett., 598, 2024
|
|
8I2K
 
 | |
4GOA
 
 | |
4G7E
 
 | |
1W3V
 
 | |
1W3X
 
 | | Isopenicillin N synthase d-(L-a-aminoadipoyl)-(3R)-methyl-L-cysteine D-a-hydroxyisovaleryl ester complex (Oxygen exposed 5 minutes 20 bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N~6~-[(1R)-1-({[(1R,2R)-1-CARBOXY-3-HYDROXY-2-METHYLPROPYL]OXY}CARBONYL)-2-MERCAPTOPROP-2-EN-1-YL]-6-OXO-L-LYSINE | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-07-20 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unexpected Oxidation of a Depsipeptide Substrate Analogue in Crystalline Isopenicillin N Synthase.
Chembiochem, 7, 2006
|
|
1CNR
 
 | |
4BB3
 
 | | Isopenicillin N synthase with the dipeptide substrate analogue AhC | | Descriptor: | (2S)-2-azanyl-6-oxidanylidene-6-[[(2S)-1-oxidanyl-1-oxidanylidene-4-sulfanyl-butan-2-yl]amino]hexanoic acid, FE (III) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2012-09-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of Isopenicillin N Synthase with a Dipeptide Substrate Analogue.
Arch.Biochem.Biophys., 530, 2013
|
|
3ZKU
 
 | | Isopenicillin N synthase with substrate analogue AhCV | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[(5S)-5-amino-5-carboxypentanoyl]-L-homocysteyl-D-valine | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Interaction of Isopenicillin N Synthase with Homologated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Homocysteinyl-D-Xaa Characterised by Protein Crystallography.
Chembiochem, 14, 2013
|
|
3ZKY
 
 | | Isopenicillin N synthase with substrate analogue AhCmC | | Descriptor: | FE (III) ION, GLYCEROL, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2013-01-25 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Interaction of Isopenicillin N Synthase with Homologated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Homocysteinyl-D-Xaa Characterised by Protein Crystallography.
Chembiochem, 14, 2013
|
|
8JI1
 
 | | Crystal structure of Ham1 from Plasmodium falciparum | | Descriptor: | Inosine triphosphate pyrophosphatase | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2023-05-25 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of nucleotide housekeeping protein HAM1 from human malaria parasite Plasmodium falciparum.
Febs J., 2024
|
|
8JHU
 
 | | Legionella effector protein SidI | | Descriptor: | Legionella pneumophila effector protein SidI | | Authors: | Wang, L, Subramanian, A, Mukherjee, S, Walter, P. | | Deposit date: | 2023-05-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A Legionella toxin exhibits tRNA mimicry and glycosyl transferase activity to target the translation machinery and trigger a ribotoxic stress response.
Nat.Cell Biol., 25, 2023
|
|
1AB1
 
 | | SI FORM CRAMBIN | | Descriptor: | CRAMBIN (SER22/ILE25), ETHANOL | | Authors: | Teeter, M.M, Yamano, A. | | Deposit date: | 1997-01-31 | | Release date: | 1997-08-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal structure of Ser-22/Ile-25 form crambin confirms solvent, side chain substate correlations.
J.Biol.Chem., 272, 1997
|
|
8GOG
 
 | | Structure of streptavidin mutant (S112Y-K121E) complexed with biotin-cyclopentadienyl-rhodium (III)(Cp*-Rh(III)) | | Descriptor: | CHLORIDE ION, GLYCEROL, RHODIUM(III) ION, ... | | Authors: | Sairaman, A, Mukherjee, P, Maiti, D, Bhaumik, P. | | Deposit date: | 2022-08-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiodivergent synthesis of isoindolones catalysed by a Rh(III)-based artificial metalloenzyme
Nat Synth, 3, 2024
|
|
7JRB
 
 | | Phospholipase D engineered mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Phospholipase D | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
7JRV
 
 | | Phospholipase D engineered mutant bound to phosphatidic acid (30 minute soak) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dibutanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
7JRW
 
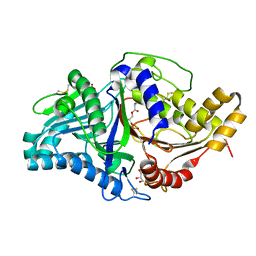 | | Phospholipase D engineered mutant bound to phosphatidic acid (5 day soak) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dibutanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
7JS5
 
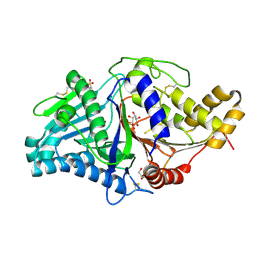 | |
7JS7
 
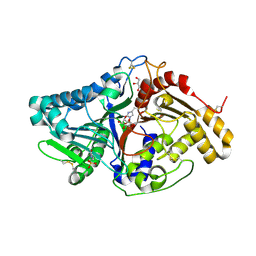 | |
7JRU
 
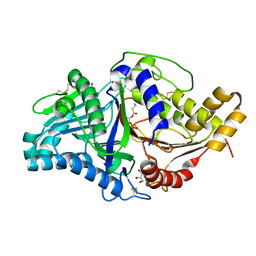 | | Phospholipase D engineered mutant bound to phosphatidic acid (8 hour soak) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dibutanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
7JRC
 
 | | Phospholipase D engineered mutant in complex with phosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
5MC6
 
 | | Cryo-EM structure of a native ribosome-Ski2-Ski3-Ski8 complex from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Schmidt, C, Kowalinski, E, Shanmuganathan, V, Defenouillere, Q, Braunger, K, Heuer, A, Pech, M, Namane, A, Berninghausen, O, Fromont-Racine, M, Jacquier, A, Conti, E, Becker, T, Beckmann, R. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex.
Science, 354, 2016
|
|
6J7W
 
 | | Crystal Structure of Human BCMA in complex with UniAb(TM) VH | | Descriptor: | Tumor necrosis factor receptor superfamily member 17, UniAb | | Authors: | Clarke, S.C, Ma, B, Trinklein, N.D, Schellenberger, U, Osborn, M, Ouisse, L, Boudreau, A, Davison, L, Harris, K.E, Ugamraj, H, Balasubramani, A, Dang, K, Jorgensen, B, Ogana, H, Pham, D, Pratap, P, Sankaran, P, Anegon, I, van Schooten, W, Bruggemann, M, Buelow, R, Force Aldred, S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multispecific Antibody Development Platform Based on Human Heavy Chain Antibodies
Front Immunol, 9, 2018
|
|
6B5V
 
 | | Structure of TRPV5 in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Lodowski, D.T, Huynh, K.W, Yazici, A, del Rosario, J, Kapoor, A, Basak, S, Samanta, A, Chakrapani, S, Zhou, Z.H, Filizola, M, Rohacs, T, Han, S, Moiseenkova-Bell, V.Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3UK6
 
 | | Crystal Structure of the Tip48 (Tip49b) hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 2 | | Authors: | Petukhov, M, Dagkessamanskaja, A, Bommer, M, Barrett, T, Tsaneva, I, Yakimov, A, Queval, R, Shvetsov, A, Khodorkovskiy, M, Kas, E, Grigoriev, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Large-Scale Conformational Flexibility Determines the Properties of AAA+ TIP49 ATPases.
Structure, 20, 2012
|
|
