4ICG
 
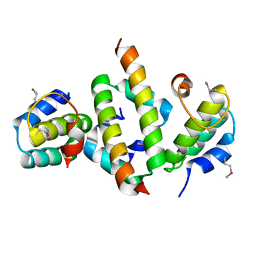 | | N-terminal dimerization domain of H-NS in complex with Hha (Salmonella Typhimurium) | | Descriptor: | DNA-binding protein H-NS, Hemolysin expression modulating protein (Involved in environmental regulation of virulence factors) | | Authors: | Ali, S.S, Whitney, J.C, Stevenson, J, Robinson, H, Howell, P.L, Navarre, W.W. | | Deposit date: | 2012-12-10 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9217 Å) | | Cite: | Structural Insights into the Regulation of Foreign Genes in Salmonella by the Hha/H-NS Complex.
J.Biol.Chem., 288, 2013
|
|
8UZ1
 
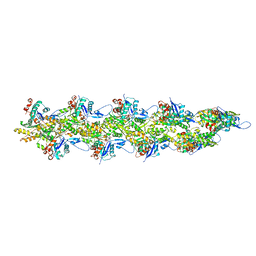 | | Straight actin filament from Arp2/3 branch junction sample (ADP-BeFx) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UXW
 
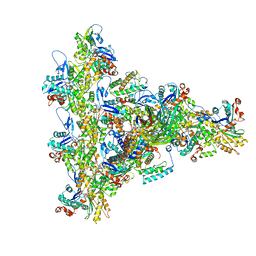 | | Arp2/3 branch junction complex, ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UXX
 
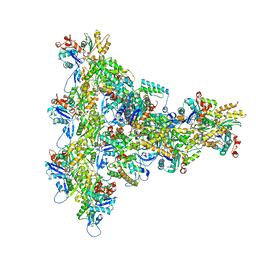 | | Arp2/3 branch junction complex, BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UZ0
 
 | | Straight actin filament from Arp2/3 branch junction sample (ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
6XH1
 
 | |
6XH0
 
 | |
6XH3
 
 | |
6XH2
 
 | |
4KWV
 
 | | Crystal Structure of human apo-QPRT | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Malik, S.S, Patterson, D.N, Ncube, Z, Toth, E.A. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid.
Proteins, 82, 2014
|
|
4KWW
 
 | | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], PHTHALIC ACID | | Authors: | Malik, S.S, Dimeka, P.N, Ncube, Z, Toth, E.A. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid.
Proteins, 82, 2014
|
|
4XEG
 
 | | Structure of the enzyme-product complex resulting from TDG action on a G/hmU mismatch | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2014-12-23 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z47
 
 | | Structure of the enzyme-product complex resulting from TDG action on a GU mismatch in the presence of excess base | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA, ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-01 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z7Z
 
 | | Structure of the enzyme-product complex resulting from TDG action on a GT mismatch in the presence of excess base | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z7B
 
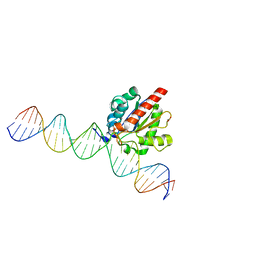 | | Structure of the enzyme-product complex resulting from TDG action on a GfC mismatch | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z3A
 
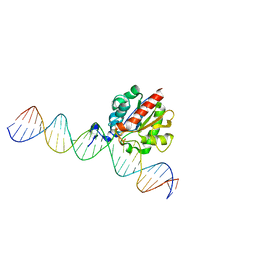 | |
5CYS
 
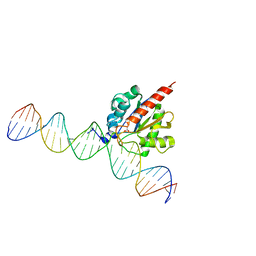 | |
5JXY
 
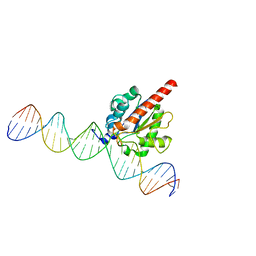 | | Enzyme-substrate complex of TDG catalytic domain bound to a G/U analog | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pidugu, L.S, Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5FF8
 
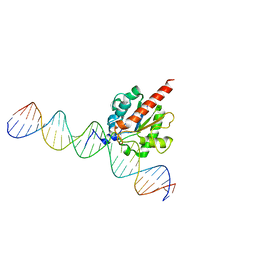 | | TDG enzyme-product complex | | Descriptor: | DNA, G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
5HF7
 
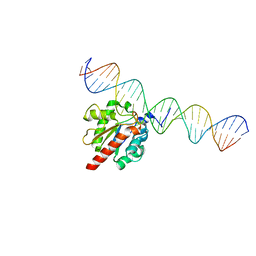 | | TDG enzyme-substrate complex | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
7KJR
 
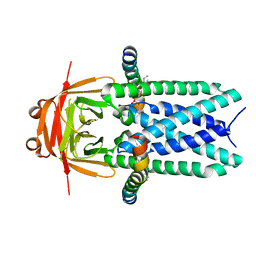 | | Cryo-EM structure of SARS-CoV-2 ORF3a | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Kern, D.M, Hoel, C.M, Kotecha, A, Brohawn, S.G. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6XDC
 
 | |
6U15
 
 | |
6U16
 
 | | Human thymine DNA glycosylase N140A mutant bound to DNA with 5-carboxyl-dC substrate | | Descriptor: | 1,2-ETHANEDIOL, DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2019-08-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Excision of 5-Carboxylcytosine by Thymine DNA Glycosylase.
J.Am.Chem.Soc., 141, 2019
|
|
6U17
 
 | | Human thymine DNA glycosylase bound to DNA with 2'-F-5-carboxyl-dC substrate analog | | Descriptor: | ACETATE ION, DNA (28-MER), DNA (30-MER), ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2019-08-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Excision of 5-Carboxylcytosine by Thymine DNA Glycosylase.
J.Am.Chem.Soc., 141, 2019
|
|
