4M8G
 
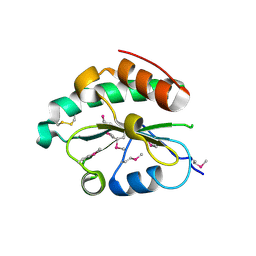 | | Crystal structure of Se-Met hN33/Tusc3 | | Descriptor: | Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-13 | | Release date: | 2014-03-26 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M90
 
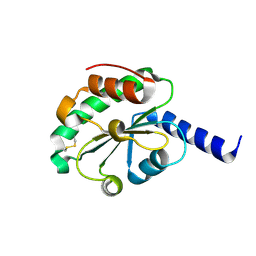 | | crystal structure of oxidized hN33/Tusc3 | | Descriptor: | Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
7OCI
 
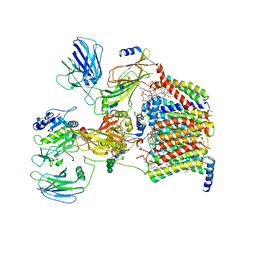 | | Cryo-EM structure of yeast Ost6p containing oligosaccharyltransferase complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Neuhaus, J.D, Eyring, J, Irobalieva, R.N, Kowal, J, Lin, C.W, Locher, K.P, Aebi, M. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Functional analysis of Ost3p and Ost6p containing yeast oligosaccharyltransferases.
Glycobiology, 31, 2021
|
|
3N0K
 
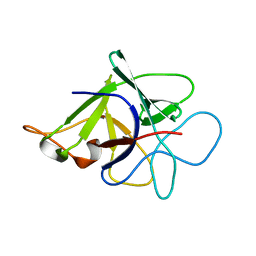 | | Proteinase inhibitor from Coprinopsis cinerea | | Descriptor: | Serine protease inhibitor 1 | | Authors: | Renko, M, Sabotic, J, Bleuler-Martinez, S, Kallert, S, Avanzo, P, Kos, J, Aebi, M, Kuenzler, M, Turk, D. | | Deposit date: | 2010-05-14 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Trypsin Inhibition and Entomotoxicity of Cospin, Serine Protease Inhibitor Involved in Defense of Coprinopsis cinerea Fruiting Bodies.
J.Biol.Chem., 287, 2012
|
|
6ZRW
 
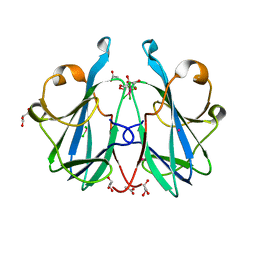 | | Crystal structure of the fungal lectin CML1 | | Descriptor: | ACETATE ION, GLYCEROL, Mucin-binding lectin 1, ... | | Authors: | Bleuler-Martinez, S, Olieric, V, Sharpe, M, Capitani, G, Aebi, M, Kuenzler, M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
3GA4
 
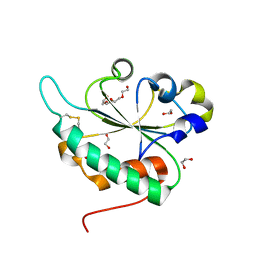 | | Crystal structure of Ost6L (photoreduced form) | | Descriptor: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2R0F
 
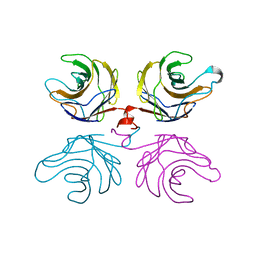 | | Ligand free structure of fungal lectin CGL3 | | Descriptor: | CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
2R0H
 
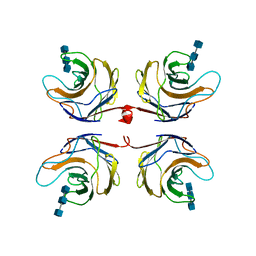 | | Fungal lectin CGL3 in complex with chitotriose (chitotetraose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CGL3 lectin | | Authors: | Waelti, M.A, Walser, P.J, Thore, S, Gruenler, A, Ban, N, Kuenzler, M, Aebi, M. | | Deposit date: | 2007-08-20 | | Release date: | 2008-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Chitotetraose Coordination by CGL3, a Novel Galectin-Related Protein from Coprinopsis cinerea
J.Mol.Biol., 379, 2008
|
|
6SNI
 
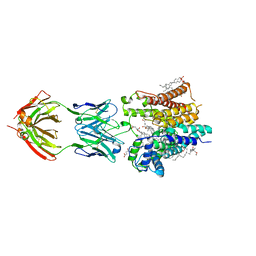 | | Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | | Descriptor: | 6AG9-Fab heavy chain, 6AG9-Fab light chain, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
6SNH
 
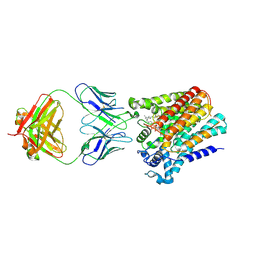 | | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc | | Descriptor: | 6AG9 Fab heavy chain, 6AG9 Fab light chain, Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
3G9B
 
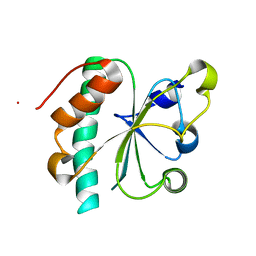 | | Crystal structure of reduced Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8AGE
 
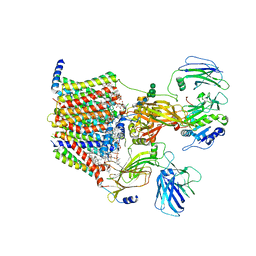 | | Structure of yeast oligosaccharylransferase complex with acceptor peptide bound | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
8AGB
 
 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide bound | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
8AGC
 
 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide and non-acceptor peptide bound | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
2K32
 
 | | Truncated AcrA from Campylobacter jejuni for glycosylation studies | | Descriptor: | A | | Authors: | Slynko, V, Schubert, M, Numao, S, Kowarik, M, Aebi, M, Allain, F. | | Deposit date: | 2008-04-17 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation.
J.Am.Chem.Soc., 131, 2009
|
|
2WKK
 
 | | Identification of the glycan target of the nematotoxic fungal galectin CGL2 in Caenorhabditis elegans | | Descriptor: | GALECTIN-2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Butschi, A, Titz, A, Waelti, M, Olieric, V, Paschinger, K, Xiaoqiang, G, Seeberger, P.H, Wilson, I.B.H, Aebi, M, Hengartner, M.O, Kuenzler, M. | | Deposit date: | 2009-06-14 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caenorhabditis Elegans N-Glycan Core Beta-Galactoside Confers Sensitivity Towards Nematotoxic Fungal Galectin Cgl2.
Plos Pathog., 6, 2010
|
|
2K33
 
 | | Solution structure of an N-glycosylated protein using in vitro glycosylation | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-[beta-D-glucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, AcrA | | Authors: | Slynko, V, Schubert, M, Numao, S, Kowarik, M, Aebi, M, Allain, F.H.-T. | | Deposit date: | 2008-04-18 | | Release date: | 2009-02-03 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation.
J.Am.Chem.Soc., 131, 2009
|
|
3G7Y
 
 | | Crystal structure of oxidized Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4M92
 
 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
 | | crystal structure of hN33/Tusc3-peptide 1 | | Descriptor: | Protein cereblon, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
3RCE
 
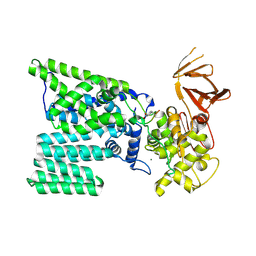 | | Bacterial oligosaccharyltransferase PglB | | Descriptor: | MAGNESIUM ION, Oligosaccharide transferase to N-glycosylate proteins, Substrate Mimic Peptide | | Authors: | Lizak, C, Gerber, S, Numao, S, Aebi, M, Locher, K.P. | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a bacterial oligosaccharyltransferase.
Nature, 474, 2011
|
|
2LIQ
 
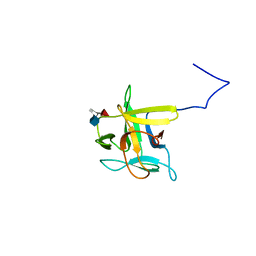 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2LIE
 
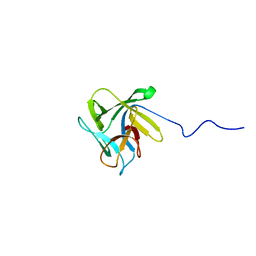 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2JJ6
 
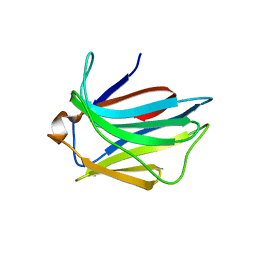 | | Crystal structure of the putative carbohydrate recognition domain of the human galectin-related protein | | Descriptor: | GALECTIN-RELATED PROTEIN | | Authors: | Thore, S, Walti, M.A, Kunzler, M, Aebi, M. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Carbohydrate Recognition Domain of Human Galectin-Related Protein
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
6EZN
 
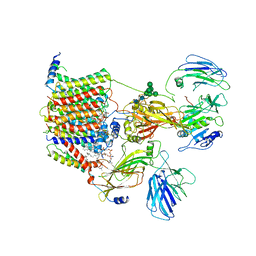 | | Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Kowal, J, Eyring, J, Ngwa, E.M, Aebi, M, Locher, K.P. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-17 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
