2PE5
 
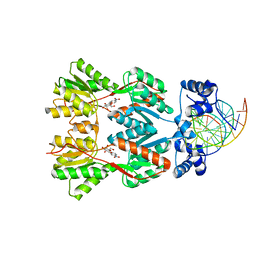 | | Crystal Structure of the Lac Repressor bound to ONPG in repressed state | | Descriptor: | 2-nitrophenyl beta-D-galactopyranoside, DNA (5'-D(*DAP*DAP*DTP*DTP*DGP*DTP*DGP*DAP*DGP*DCP*DGP*DCP*DTP*DCP*DAP*DCP*DAP*DAP*DTP*DT)-3'), Lactose operon repressor | | Authors: | Daber, R, Stayrook, S.E, Rosenberg, A, Lewis, M. | | Deposit date: | 2007-04-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of lac repressor bound to allosteric effectors
J.Mol.Biol., 370, 2007
|
|
2PAF
 
 | |
2P9H
 
 | |
3N73
 
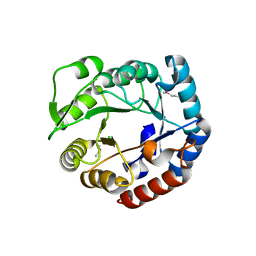 | | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus | | Descriptor: | CHLORIDE ION, Putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Cabello, R, Chruszcz, M, Xu, X, Zimmerman, M.D, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus
To be Published
|
|
6AAV
 
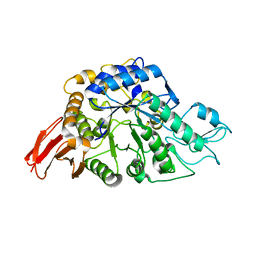 | | Crystal structure of alpha-glucosyl transfer enzyme, XgtA at 1.72 angstrom resolution | | Descriptor: | Alpha-glucosyltransferase | | Authors: | Kurumizaka, H, Arimura, Y, Kirimura, K, Watanabe, R. | | Deposit date: | 2018-07-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of alpha-glucosyl transfer enzyme XgtA from Xanthomonas campestris WU-9701.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
7CIO
 
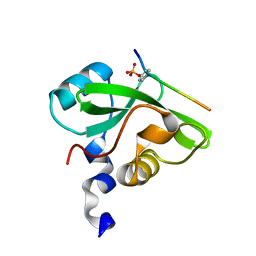 | | Molecular interactions of cytoplasmic region of CTLA-4 with SH2 domains of PI3-kinase | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Iiyama, M, Numoto, N, Ogawa, S, Kuroda, M, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular interactions of the CTLA-4 cytoplasmic region with the phosphoinositide 3-kinase SH2 domains.
Mol.Immunol., 131, 2021
|
|
5GJI
 
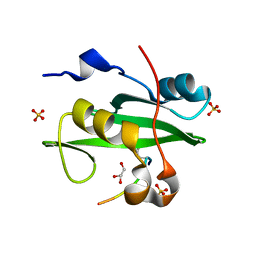 | | PI3K p85 N-terminal SH2 domain/CD28-derived peptide complex | | Descriptor: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION, ... | | Authors: | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-14 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
5GJH
 
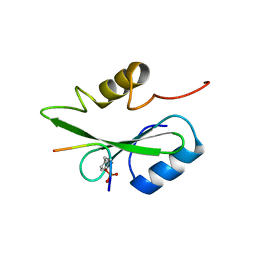 | | Gads SH2 domain/CD28-derived peptide complex | | Descriptor: | GRB2-related adapter protein 2, T-cell-specific surface glycoprotein CD28 | | Authors: | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
6ICH
 
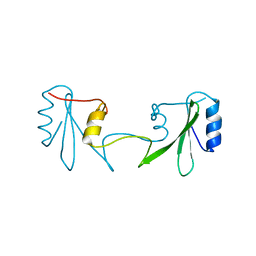 | | Grb2 SH2 domain in domain swapped dimer form | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
6ICG
 
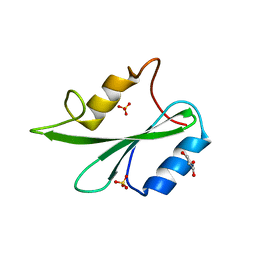 | | Grb2 SH2 domain in phosphopeptide free form | | Descriptor: | GLYCEROL, Growth factor receptor-bound protein 2, SULFATE ION | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
8S99
 
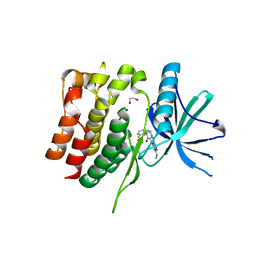 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 11 | | Descriptor: | (8S)-N-[(1R,2S)-2-fluorocyclopropyl]-5-{[(1M,2'M)-3'-fluoro-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S9A
 
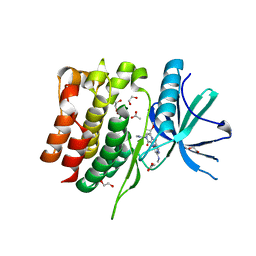 | | Crystal structure of the TYK2 pseudokinase domain in complex with TAK-279 | | Descriptor: | (8S)-N-[(1R,2R)-2-methoxycyclobutyl]-7-(methylamino)-5-{[(1P,2'P)-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S98
 
 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 8 | | Descriptor: | (8S)-N-cyclopropyl-5-[(2-methoxypyridin-3-yl)amino]-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
6DD3
 
 | | Crystal structure of the double mutant (D52N/D407A) of NT5C2-537X in the active state | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDC
 
 | | Crystal structure of the single mutant (D52N) of NT5C2-537X in the basal state, Northeast Structural Genomics Consortium Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DE3
 
 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-11 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDZ
 
 | | Crystal structure of the double mutant (D52N/R238W) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDO
 
 | | Crystal structure of the single mutant (D52N) of the full-length NT5C2 in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDB
 
 | | Crystal structure of the double mutant (D52N/R367Q) of NT5C2-537X in the basal state, Northeast Structural Genomics Consortium Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDY
 
 | | Crystal structure of the double mutant (D52N/K359Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDX
 
 | | Crystal structure of the double mutant (D52N/L375F) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1G
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from 50,000 diffraction patterns. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
