5JQS
 
 | |
5JKN
 
 | | Crystal structure of deubiquitinase MINDY-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MERCURY (II) ION, PHOSPHATE ION, ... | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes.
Mol.Cell, 63, 2016
|
|
6Y6R
 
 | | Crystal structure of MINDY1 T335D mutant | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY-1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6YJG
 
 | | Crystal structure of MINDY1 mutant-Y114F | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6Z49
 
 | | Crystal structure of deubiquitinase Mindy2 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-05-23 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6Z7V
 
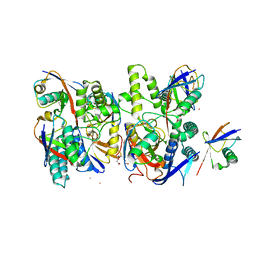 | | Crystal structure of Mindy2 (C266A) in complex with Lys48 linked di-ubiquitin (K48-Ub2) | | Descriptor: | POTASSIUM ION, Polyubiquitin-C, TETRAETHYLENE GLYCOL, ... | | Authors: | Abdul Rehman, S.A, Lange, S.M, Kulathu, Y. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6Z90
 
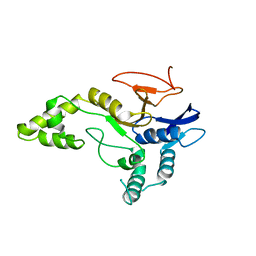 | | Crystal structure of MINDY1 mutant-P138A | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY-1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
4EHS
 
 | |
6TUV
 
 | |
6TXB
 
 | |
4IM9
 
 | |
5XW3
 
 | |
4ZGL
 
 | | Hit Like Protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Uncharacterized HIT-like protein HP_0404 | | Authors: | Tarique, K.F, Devi, S, Abdul Rehman, S.A, Gourinath, S. | | Deposit date: | 2015-04-23 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of HINT from Helicobacter pylori.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4ZG5
 
 | | Structural and functional insights into Survival endonuclease, an important virulence factor of Brucella abortus | | Descriptor: | 5'-nucleotidase SurE, MAGNESIUM ION | | Authors: | Tarique, K.F, Abdul Rehman, S.A, Devi, S, Gourinath, S. | | Deposit date: | 2015-04-22 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insights into the stationary-phase survival protein SurE, an important virulence factor of Brucella abortus
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4H7O
 
 | |
4QEZ
 
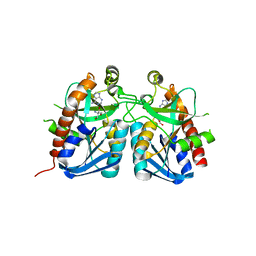 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Bacillus anthracis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE | | Authors: | Tarique, K.F, Devi, S, Abdul Rehman, S.A, Gourinath, S. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Bacillus anthracis
To be Published
|
|
4QXD
 
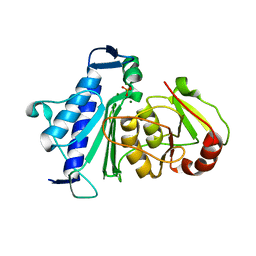 | | Crystal structure of Inositol Polyphosphate 1-Phosphatase from Entamoeba histolytica | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, putative, MAGNESIUM ION, ... | | Authors: | Tarique, K.F, Abdul Rehman, S.A, Betzel, C, Gourinath, S. | | Deposit date: | 2014-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-based identification of inositol polyphosphate 1-phosphatase from Entamoeba histolytica
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4S1Z
 
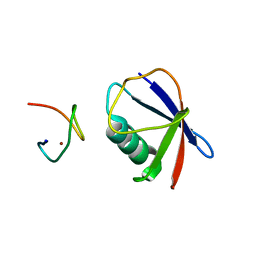 | | Crystal structure of TRABID NZF1 in complex with K29 linked di-Ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin thioesterase ZRANB1, ZINC ION | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
4S22
 
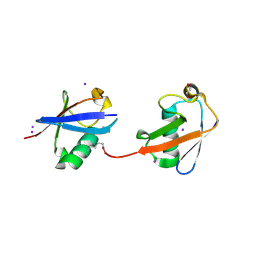 | | Crystal structure of K29 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-17 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
6FGE
 
 | | Crystal structure of human ZUFSP/ZUP1 in complex with ubiquitin | | Descriptor: | ACETATE ION, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kwasna, D, Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-04 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability.
Mol. Cell, 70, 2018
|
|
5MN9
 
 | |
7NPI
 
 | | Crystal structure of Mindy2 (C266A) in complex with Lys48-linked penta-ubiquitin (K48-Ub5) | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, SODIUM ION, ... | | Authors: | Lange, S.M, Armstrong, L.A, Kulathu, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6AHI
 
 | |
4Y1H
 
 | | Crystal structure of K33 linked tri-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
4XYZ
 
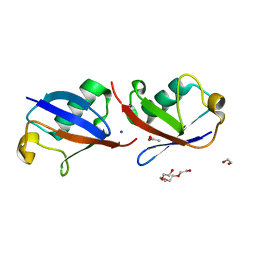 | | Crystal structure of K33 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
