4QEI
 
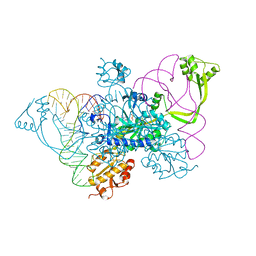 | | Two distinct conformational states of GlyRS captured in crystal lattice | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine--tRNA ligase, tRNA-Gly-CCC-2-2 | | Authors: | Xie, W, Qin, X, Deng, X, Zhang, Q, Li, Q. | | Deposit date: | 2014-05-16 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
8X96
 
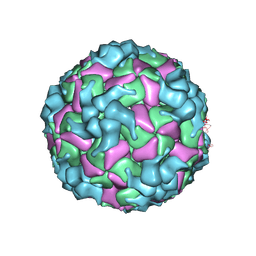 | | Cryo-EM structure of enterovirus A71 A-particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8X9B
 
 | | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2 (local refinement) | | Descriptor: | Capsid protein VP1, Genome polyprotein, The heavy chain of Fab h1A6.2, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8X98
 
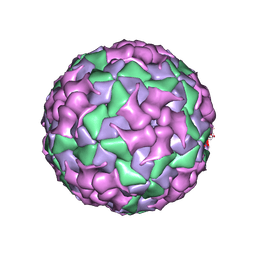 | | Cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8X99
 
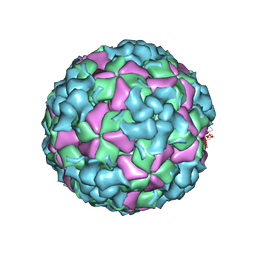 | | Cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8X95
 
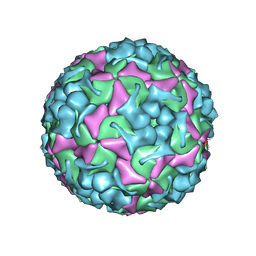 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8X9A
 
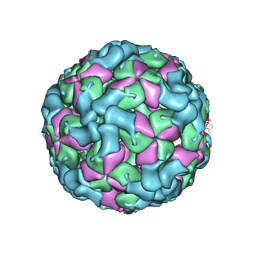 | | Cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8X97
 
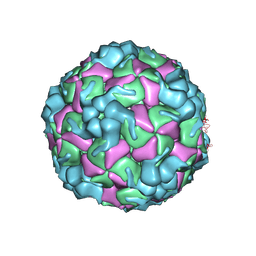 | | Cryo-EM structure of enterovirus A71 empty particle in complex with Fab h1A6.2 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8YTJ
 
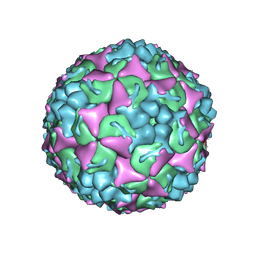 | | Cryo-EM structure of enterovirus A71 mature virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
8YTB
 
 | | Cryo-EM structure of enterovirus A71 empty particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-25 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Broadly therapeutic antibody provides cross-serotype protection against enteroviruses via Fc effector functions and by mimicking SCARB2.
Nat Microbiol, 9, 2024
|
|
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X42
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP0, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2O
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 2E6 (CVB1-M:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X49
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2I
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 2E6 (CVB1-pre-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3C
 
 | | Cryo-EM structure of Coxsackievirus B1 muture virion in complex with nAbs 8A10 and 5F5 (CVB1-M:8A10:5F5) | | Descriptor: | 5F5 heavy chain, 5F5 light chain, 8A10 heavy chain, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2W
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 8A10 (CVB1-pre-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2T
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (CVB1-M:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3E
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 9A3 (CVB1-pre-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3D
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 9A3 (CVB1-M:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X40
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
