8J3O
 
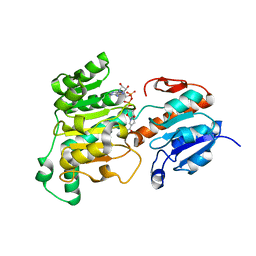 | | Formate dehydrogenase wild-type enzyme from Candida dubliniensis complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Formate dehydrogenase, MAGNESIUM ION | | Authors: | Ma, W, Zheng, Y.C, Geng, Q, Chen, C. | | Deposit date: | 2023-04-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
8J3P
 
 | | Formate dehydrogenase mutant from from Candida dubliniensis M4 complexed with NADP+ | | Descriptor: | Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ma, W, Zheng, Y.C, Geng, Q, Chen, C, Xu, J.H. | | Deposit date: | 2023-04-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
8JO2
 
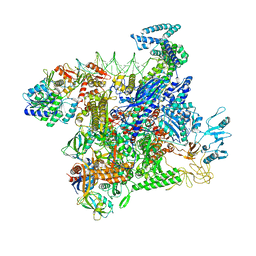 | | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA | | Descriptor: | DNA (65-MER), DNA-binding transcriptional regulator BasR, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lou, Y.-C, Huang, H.-Y, Chen, C, Wu, K.-P. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA.
Nucleic Acids Res., 51, 2023
|
|
6JK2
 
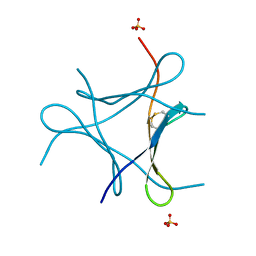 | | Crystal structure of a mini fungal lectin, PhoSL | | Descriptor: | Lectin, SULFATE ION | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
8JTD
 
 | | BJOX2000.664 trimer in complex with Fab fragment of broadly neutralizing HIV antibody PGT145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PGT145 antibody fragment, ... | | Authors: | Chatterjee, A, Chen, C, Lee, K, Mangala Prasad, V. | | Deposit date: | 2023-06-21 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | An HIV-1 broadly neutralizing antibody overcomes structural and dynamic variation through highly focused epitope targeting.
Npj Viruses, 1, 2023
|
|
8JTM
 
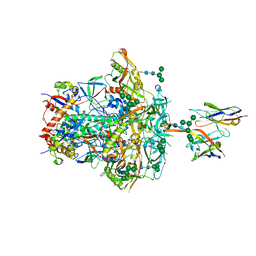 | | CNE55.664 trimer in complex with broadly neutralizing HIV antibody PGT145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PGT145 antibody fragment, ... | | Authors: | Chatterjee, A, Chen, C, Lee, K, Mangala Prasad, V. | | Deposit date: | 2023-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | An HIV-1 broadly neutralizing antibody overcomes structural and dynamic variation through highly focused epitope targeting.
Npj Viruses, 1, 2023
|
|
6IDO
 
 | | Crystal structure of Klebsiella pneumoniae sigma4 of sigmaS fusing with the RNA polymerase beta-flap-tip-helix in complex with -35 element DNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*AP*TP*TP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*C)-3'), RNA polymerase sigma factor RpoS,RNA polymerase beta-flap-tip-helix | | Authors: | Lou, Y.C, Chien, C.Y, Chen, C, Hsu, C.H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.748 Å) | | Cite: | Structural basis for -35 element recognition by sigma4chimera proteins and their interactions with PmrA response regulator.
Proteins, 88, 2020
|
|
6JK3
 
 | | Crystal structure of a mini fungal lectin, PhoSL in complex with core-fucosylated chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lectin | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
7VE6
 
 | | N-terminal domain of VraR | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator protein VraR | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE4
 
 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE5
 
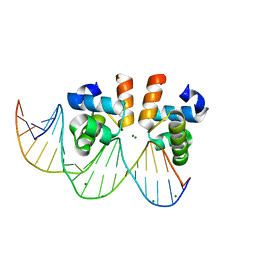 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator, MAGNESIUM ION, R1-DNA | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7XT0
 
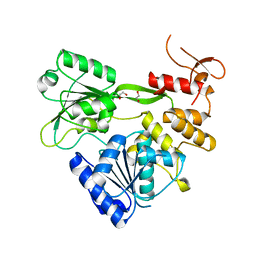 | | Crystal structure of RNA helicase from Saint Louis encephalitis virus and discovery of its inhibitors | | Descriptor: | 1,2-ETHANEDIOL, RNA helicase | | Authors: | Wang, D.P, Jiang, F.Y, Zeng, X.Y, Zhao, R, Chen, C, Zhu, Y, Cao, J.M. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of RNA helicase from Saint Louis encephalitis virus and discovery of its inhibitors.
Genes Dis, 10, 2023
|
|
7CWX
 
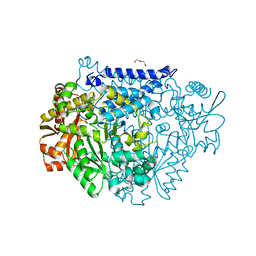 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Decarboxylase, GLYCEROL | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis
to be published
|
|
7CX0
 
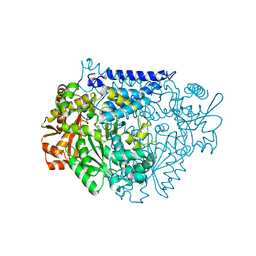 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa | | Descriptor: | CARBIDOPA, Decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa
to be published
|
|
7CWY
 
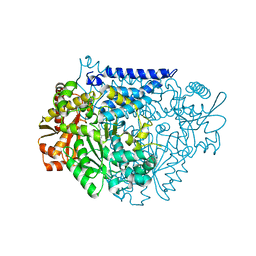 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP | | Descriptor: | Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP
to be published
|
|
7CX1
 
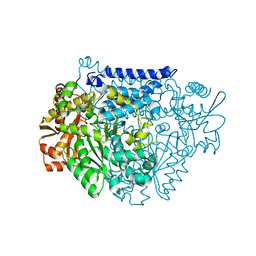 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine | | Descriptor: | 4-[(2R)-2-(methylamino)propyl]phenol, Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine
to be published
|
|
7CWZ
 
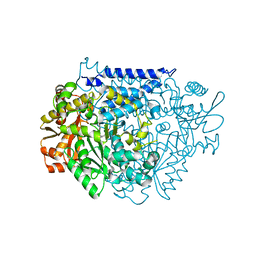 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa | | Descriptor: | Decarboxylase, L-DOPAMINE, MAGNESIUM ION, ... | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa
to be published
|
|
7BQA
 
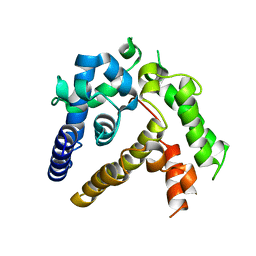 | | Crystal structure of ASFV p35 | | Descriptor: | 60 kDa polyprotein | | Authors: | Li, G.B, Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
7E58
 
 | | interferon-inducible anti-viral protein 2 | | Descriptor: | Guanylate-binding protein 2 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E5A
 
 | | interferon-inducible anti-viral protein R356A | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E59
 
 | | interferon-inducible anti-viral protein truncated | | Descriptor: | Guanylate-binding protein 5 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OS9
 
 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant with purification tag | | Descriptor: | IMIDAZOLE, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Lo Leggio, L, Von Wachenfeldt, C, Carey, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7ABX
 
 | | Perdeuterated E65Q-TIM complexed with 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
7AZA
 
 | | Perdeuterated E65Q-TIM complexed with PHOSPHOGLYCOLOHYDROXAMATE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-11-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
7AZ4
 
 | | Perdeuterated E65Q-TIM complexed with 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kelpsas, V, Caldararu, O, von Wachenfeldt, C, Oksanen, E. | | Deposit date: | 2020-11-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.15 Å), X-RAY DIFFRACTION | | Cite: | Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps.
Iucrj, 8, 2021
|
|
