8CQC
 
 | |
4ZA5
 
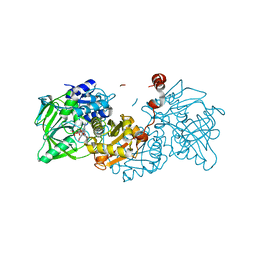 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium and ketimine forms. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, Fdc1, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
3DHC
 
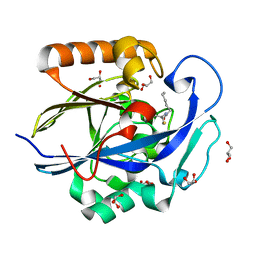 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
7WVM
 
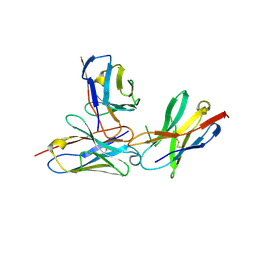 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
5HRY
 
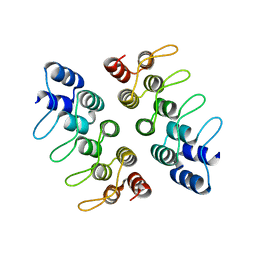 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
1FZV
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PLACENTA GROWTH FACTOR-1 (PLGF-1), AN ANGIOGENIC PROTEIN AT 2.0A RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PLACENTA GROWTH FACTOR | | Authors: | Iyer, S, Leonidas, D.D, Swaminathan, G.J, Maglione, D, Battisti, M, Tucci, M, Persico, M.G, Acharya, K.R. | | Deposit date: | 2000-10-04 | | Release date: | 2001-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution.
J.Biol.Chem., 276, 2001
|
|
8OKI
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex bound to Spt4/5 | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
4ZA4
 
 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAF
 
 | |
4ZAY
 
 | | Structure of UbiX E49Q in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
3ZE5
 
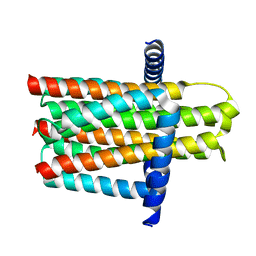 | | Crystal structure of the integral membrane diacylglycerol kinase - delta4 | | Descriptor: | DIACYLGLYCEROL KINASE | | Authors: | Li, D, Vogeley, L, Pye, V.E, Lyons, J.A, Aragao, D, Caffrey, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
3ZE3
 
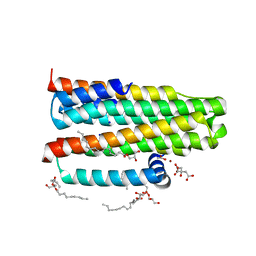 | | Crystal structure of the integral membrane diacylglycerol kinase - delta7 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Li, D, Pye, V.E, Lyons, J.A, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
3ZE4
 
 | | Crystal structure of the integral membrane diacylglycerol kinase - wild-type | | Descriptor: | DIACYLGLYCEROL KINASE | | Authors: | Li, D, Lyons, J.A, Pye, V.E, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.702 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
6RQO
 
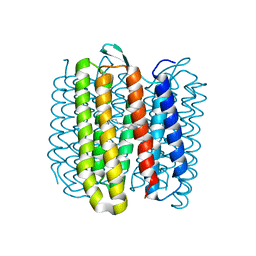 | | Steady-state-SMX activated state structure of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
5Z3G
 
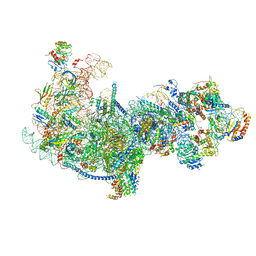 | | Cryo-EM structure of a nucleolar pre-60S ribosome (Rpf1-TAP) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Zhu, X, Zhou, D, Ye, K. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate.
Protein Cell, 10, 2019
|
|
4ZAG
 
 | |
5YZC
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor compound (AS-48) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitro-2-[(phenylacetyl)amino]benzamide, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZD
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor peptide (FIP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8JSW
 
 | | Human VMAT2 complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
3DHA
 
 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
8JTC
 
 | | Human VMAT2 complex with reserpine | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
5JXV
 
 | | Solid-state MAS NMR structure of immunoglobulin beta 1 binding domain of protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Andreas, L.B, Jaudzems, K, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JZR
 
 | | Solid-state MAS NMR structure of Acinetobacter phage 205 (AP205) coat protein in assembled capsid particles | | Descriptor: | Coat protein | | Authors: | Jaudzems, K, Andreas, L.B, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8K0B
 
 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
8JT9
 
 | | Human VMAT2 complex with ketanserin | | Descriptor: | 3-[2-[4-(4-fluorophenyl)carbonylpiperidin-1-yl]ethyl]-1~{H}-quinazoline-2,4-dione, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
