4V3L
 
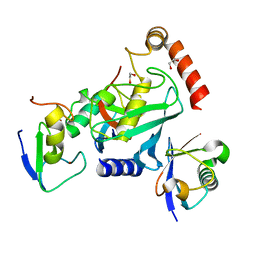 | | RNF38-UB-UbcH5B-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF38, POLYUBIQUITIN-C, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W1V
 
 | | The beta2 adrenergic receptor bound to a bitopic ligand | | Descriptor: | (2S)-1-[(3-{1-[4-(4-{(2S)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}phenyl)butyl]-1H-1,2,3-triazol-4-yl}propyl)amino]-3-(2-propylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Endolysin, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Gaiser, B, Danielsen, M, Xu, X, Jorgensen, K, Fronik, P, Marcher-Rorsted, E, Wrobe, T, Hirata, K, Liu, X, Mathiesen, J, Pedersen, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bitopic Ligands Support the Presence of a Metastable Binding Site at the beta 2 Adrenergic Receptor.
J.Med.Chem., 67, 2024
|
|
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
3KLS
 
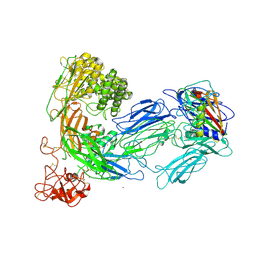 | | Structure of complement C5 in complex with SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3N23
 
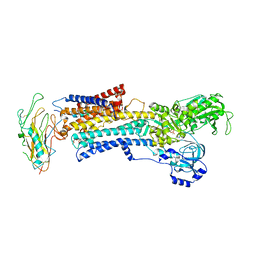 | | Crystal structure of the high affinity complex between ouabain and the E2P form of the sodium-potassium pump | | Descriptor: | MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, OUABAIN, ... | | Authors: | Yatime, L, Laursen, M, Morth, J.P, Esmann, M, Nissen, P, Fedosova, N.U. | | Deposit date: | 2010-05-17 | | Release date: | 2011-01-19 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase.
J.Struct.Biol., 174, 2011
|
|
7EXZ
 
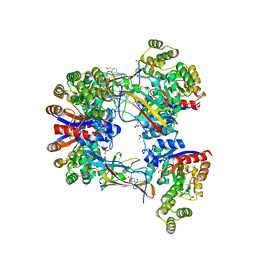 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
 | | DfgA-DfgB complex apo 2.4 angstrom | | Descriptor: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7MF0
 
 | | Co-crystal structure of PERK with inhibitor (R)-2-amino-N-cyclopropyl-5-(4-(2-(3,5-difluorophenyl)-2-hydroxyacetamido)-2-methylphenyl)nicotinamide | | Descriptor: | 2-amino-N-cyclopropyl-5-(4-{[(2R)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino}-2-methylphenyl)pyridine-3-carboxamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wiens, B, Koszelak-Rosenblum, M, Surman, M.D, Zhu, G, Mulvihill, M.J. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors.
Bioorg.Med.Chem.Lett., 43, 2021
|
|
5LXI
 
 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
5LXH
 
 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | Cysteine protease ATG4B, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
4R9P
 
 | | An Expansion to the Smad MH2-family: The structure of the N-MH2 expanded domain | | Descriptor: | RE28239p | | Authors: | Beich-Frandsen, M, Aragon, E, Llimargas, M, Benach, J, Riera, A, Macias, M.J. | | Deposit date: | 2014-09-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Structure of the N-terminal domain of the protein Expansion: an 'Expansion' to the Smad MH2
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8JW3
 
 | | The crystal structure of the viral terpene synthase from Orpheovirus IHUMI-LCC2 | | Descriptor: | SULFATE ION, Terpenoid synthase | | Authors: | Jung, Y, Mitsuhashi, T, Senda, M, Sato, S, Senda, T, Fujita, M. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Function and Structure of a Terpene Synthase Encoded in a Giant Virus Genome.
J.Am.Chem.Soc., 145, 2023
|
|
4F4O
 
 | | Structure of the Haptoglobin-Haemoglobin Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Andersen, C.B.F, Torvund-Jensen, M, Nielsen, M.J, Oliveira, C.L.P, Hersleth, H.P, Andersen, N.H, Pedersen, J.S, Andersen, G.R, Moestrup, S.K. | | Deposit date: | 2012-05-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the haptoglobin-haemoglobin complex.
Nature, 489, 2012
|
|
6WO0
 
 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | Descriptor: | GLYCEROL, Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
6WNL
 
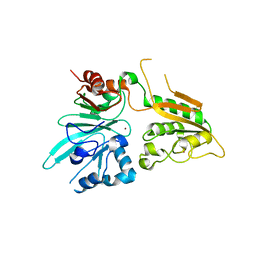 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | Descriptor: | Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
8OM9
 
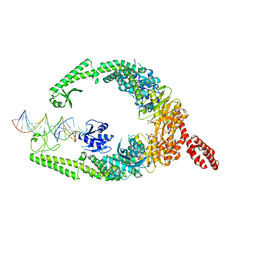 | | MutSbeta bound to (CAG)2 DNA (open form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (open form)
To Be Published
|
|
8OLX
 
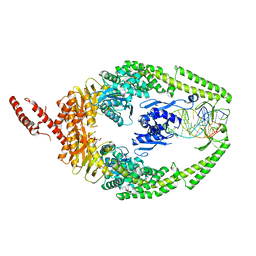 | | MutSbeta bound to (CAG)2 DNA (canonical form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein Msh2, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (canonical form)
To Be Published
|
|
8OMO
 
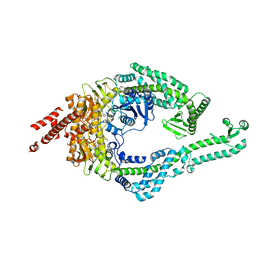 | | DNA-unbound MutSbeta-ATP complex (bent clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (bent clamp form)
To Be Published
|
|
8OM5
 
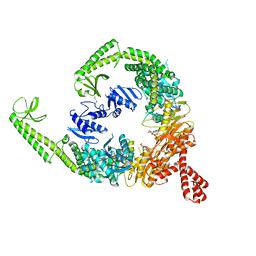 | | DNA-free open form of MutSbeta | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-free open form of MutSbeta
To Be Published
|
|
8OMQ
 
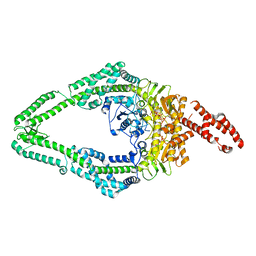 | | DNA-unbound MutSbeta-ATP complex (straight clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (straight clamp form)
To Be Published
|
|
8OMA
 
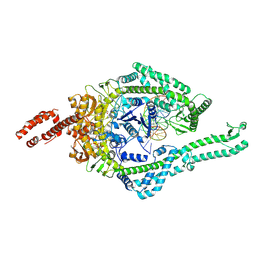 | | MutSbeta bound to 61bp homoduplex DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | MutSbeta bound to 61bp homoduplex DNA
To Be Published
|
|
6VR0
 
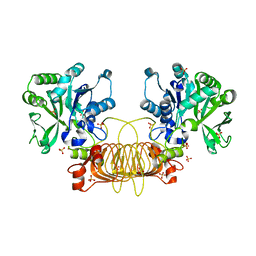 | | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Liu, D, Ballicora, M, Iglesias, A, Asencion, M, Figueroa, C. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A
To Be Published
|
|
5XNZ
 
 | | Crystal structure of CreD complex with fumarate | | Descriptor: | CreD, FUMARIC ACID | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
