7TE5
 
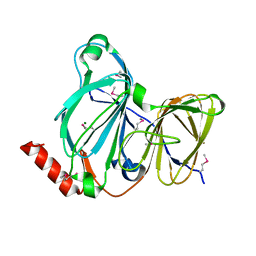 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis | | Descriptor: | MAGNESIUM ION, Pirin family protein Yhak | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis
To Be Published
|
|
7TEM
 
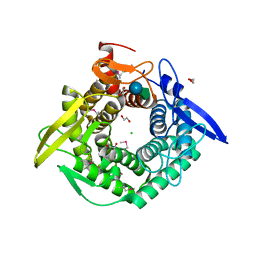 | | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-05 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis
To Be Published
|
|
7TG5
 
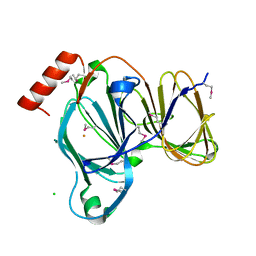 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the Presence of Fe Ion from Yersinia pestis | | Descriptor: | CHLORIDE ION, FE (III) ION, Pirin family protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the presence of Fe ion from Yersinia pestis
To Be Published
|
|
7TFQ
 
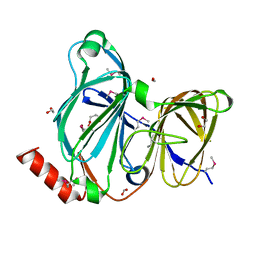 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, FORMIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis
To Be Published
|
|
2KW4
 
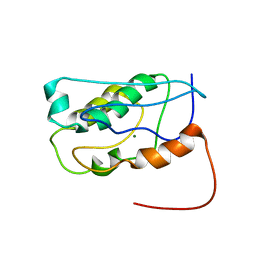 | | Solution NMR Structure of the Holo Form of a Ribonuclease H domain from D.hafniense, Northeast Structural Genomics Consortium Target DhR1A | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target DhR1A
To be Published
|
|
7STU
 
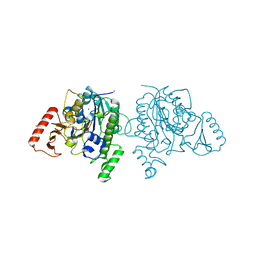 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | BROMIDE ION, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
7STT
 
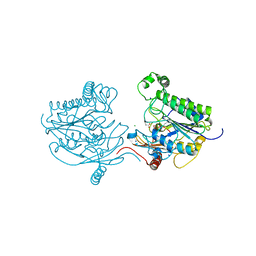 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MALONATE ION, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
2KYD
 
 | | RDC and RCSA refinement of an A-form RNA: Improvements in Major Groove Width | | Descriptor: | RNA (5'-R(*CP*UP*AP*GP*UP*UP*AP*GP*CP*UP*AP*AP*CP*UP*AP*G)-3') | | Authors: | Tolbert, B.S, Summers, M.F, Miyazaki, Y, Barton, S, Kinde, B, Stark, P, Singh, R, Bax, A, Case, D. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement.
J.Biomol.Nmr, 47, 2010
|
|
2KZ7
 
 | |
7STV
 
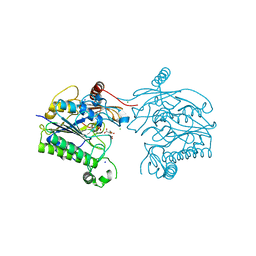 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
2KQ6
 
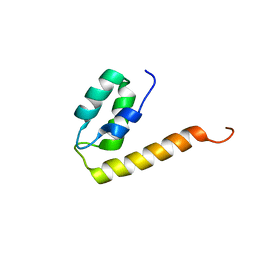 | | The structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity | | Descriptor: | Polycystin-2 | | Authors: | Petri, E.T, Celic, A, Kennedy, S.D, Ehrlich, B.E, Boggon, T.J, Hodsdon, M.E. | | Deposit date: | 2009-10-28 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the EF-hand domain of polycystin-2 suggests a mechanism for Ca2+-dependent regulation of polycystin-2 channel activity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4X90
 
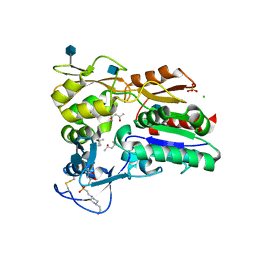 | | Crystal structure of Lysosomal Phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and function of lysosomal phospholipase A2 and lecithin:cholesterol acyltransferase.
Nat Commun, 6, 2015
|
|
2KO0
 
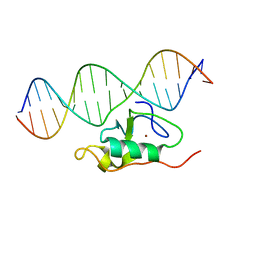 | | Solution structure of the THAP zinc finger of THAP1 in complex with its DNA target | | Descriptor: | RRM1, THAP domain-containing protein 1, ZINC ION | | Authors: | Campagne, S, Gervais, V, Saurel, O, Milon, A. | | Deposit date: | 2009-09-08 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of specific DNA-recognition by the THAP zinc finger
Nucleic Acids Res., 2010
|
|
4X94
 
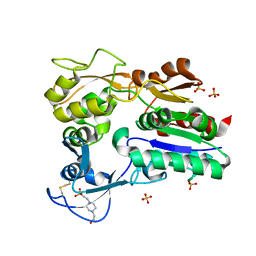 | |
4WFB
 
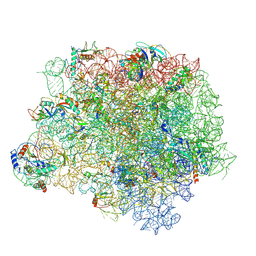 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with BC-3205 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2KSM
 
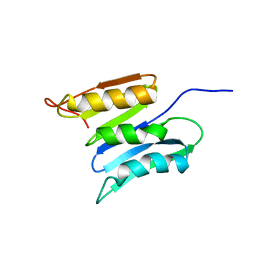 | | Central B domain of Rv0899 from Mycobacterium tuberculosis | | Descriptor: | MYCOBACTERIUM TUBERCULOSIS RV0899/MT0922/OmpATb | | Authors: | Teriete, P, Yao, Y, Kolodzik, A, Yu, J, Song, H, Niederweis, M, Marassi, F.M. | | Deposit date: | 2010-01-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mycobacterium tuberculosis Rv0899 adopts a mixed alpha/beta-structure and does not form a transmembrane beta-barrel.
Biochemistry, 49, 2010
|
|
7TFM
 
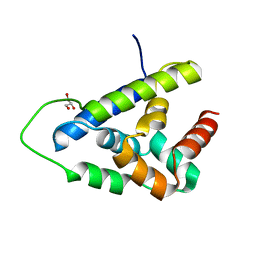 | | Atomic Structure of the Leishmania spp. Hsp100 N-Domain | | Descriptor: | ATP-dependent Clp protease subunit, heat shock protein 100 (HSP100), GLYCEROL | | Authors: | Mercado, J.M, Lee, S, Chang, C, Sung, N, Soong, L, Catic, A, Tsai, F.T.F. | | Deposit date: | 2022-01-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.055 Å) | | Cite: | Atomic structure of the Leishmania spp. Hsp100 N-domain.
Proteins, 90, 2022
|
|
4WF9
 
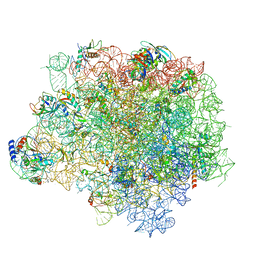 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with telithromycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.427 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2KK2
 
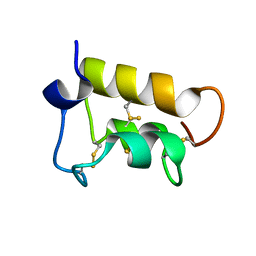 | | NMR solution structure of the pheromone En-A1 from Euplotes nobilii | | Descriptor: | En-A1 | | Authors: | Pedrini, B, Alimenti, C, Vallesi, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antarctic and Arctic populations of the ciliate Euplotes nobilii show common pheromone-mediated cell-cell signaling and cross-mating.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2KWA
 
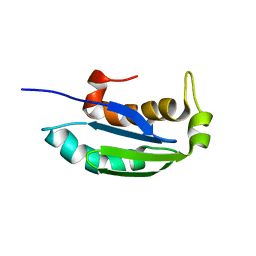 | | 1H, 13C and 15N backbone and side chain resonance assignments of the N-terminal domain of the histidine kinase inhibitor KipI from Bacillus subtilis | | Descriptor: | Kinase A inhibitor | | Authors: | Hynson, R.M.G, Kwan, A, Jacques, D.A, Mackay, J.P, Trewhella, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel Structure of an Antikinase and its Inhibitor
J.Mol.Biol., 2010
|
|
2KYZ
 
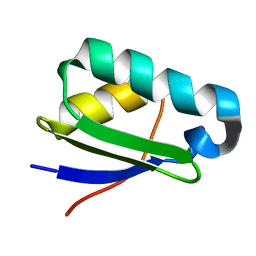 | | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima | | Descriptor: | Heavy metal binding protein | | Authors: | Jaudzems, K, Wahab, A, Serrano, P, Geralt, M, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima
To be Published
|
|
2KM3
 
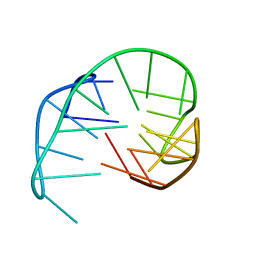 | | Structure of an intramolecular G-quadruplex containing a G.C.G.C tetrad formed by human telomeric variant CTAGGG repeats | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*TP*AP*GP*GP*GP*CP*TP*AP*GP*GP*GP*CP*TP*AP*GP*GP*G)-3') | | Authors: | Lim, K.W, Alberti, P, Guedin, A, Lacroix, L, Riou, J.F, Royle, N.J, Mergny, J.L, Phan, A.T. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad
Nucleic Acids Res., 37, 2009
|
|
2KPQ
 
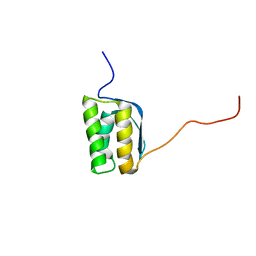 | | NMR Structure of Agrobacterium tumefaciens protein Atu1219: Northeast Structural Genomics Consortium target AtT14 | | Descriptor: | Uncharacterized protein | | Authors: | Cort, J.R, Yee, A, Arrowsmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Agrobacterium tumefaciens protein Atu1219
To be Published
|
|
2KMX
 
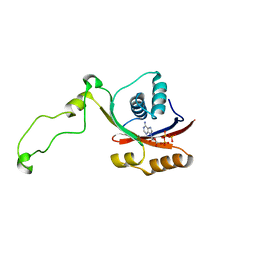 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
2KPB
 
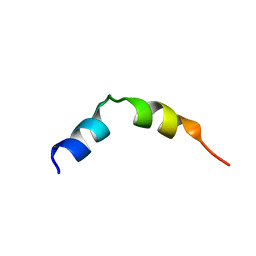 | | Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO | | Descriptor: | ARNO-p(375-400) | | Authors: | Merkulova, M, Bakulina, A, Thaker, Y.R, Gr ber, G, Marshansky, V. | | Deposit date: | 2009-10-11 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Specific motifs of the V-ATPase a2-subunit isoform interact with catalytic and regulatory domains of ARNO
Biochim.Biophys.Acta, 2010
|
|
