8X8A
 
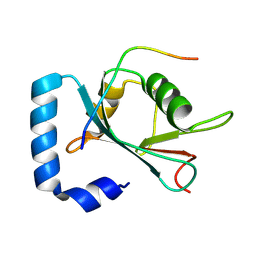 | | Crystal structure of STBD1 LIR motif in complex with GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, Starch-binding domain-containing protein 1 | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X8K
 
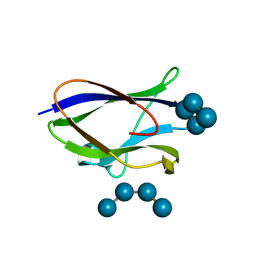 | | Crystal structure of STBD1 CBM20 domain in complex with maltotetraose | | Descriptor: | GLYCEROL, Starch-binding domain-containing protein 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8DIQ
 
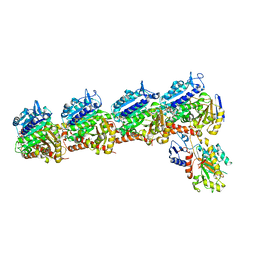 | | Tubulin-RB3_SLD-TTL in complex with SB226 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | SB226, an inhibitor of tubulin polymerization, inhibits paclitaxel-resistant melanoma growth and spontaneous metastasis.
Cancer Lett., 555, 2022
|
|
5OQ8
 
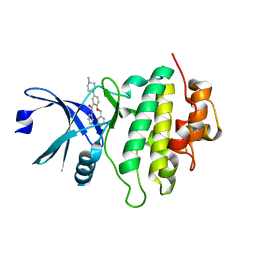 | | Structure of CHK1 12-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPU
 
 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyridine LRRK2 inhibitor | | Descriptor: | 6-azanyl-4-(3-methylphenyl)-1~{H}-pyrrolo[2,3-b]pyridine-3-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ7
 
 | | Structure of CHK1 8-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPR
 
 | | Structure of CHK1 10-pt. mutant complex with aminopyridine LRRK2 inhibitor | | Descriptor: | 5-[4-(morpholin-4-ylmethyl)phenyl]-3-(1-propan-2-yl-1,2,3-triazol-4-yl)pyridin-2-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPV
 
 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyridine LRRK2 inhibitor | | Descriptor: | 4-(3-methylphenyl)-6-[(1-methylpyrazol-3-yl)amino]-1~{H}-pyrrolo[2,3-b]pyridine-3-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ5
 
 | | Structure of CHK1 8-pt. mutant complex with aminopyrimido-benzodiazepinone LRRK2 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7DLY
 
 | | Crystal structure of Arabidopsis ACS7 mutant in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7 | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7DLW
 
 | | Crystal structure of Arabidopsis ACS7 in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7, SULFATE ION | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7U4E
 
 | |
7U4F
 
 | |
7U4G
 
 | | Neuraminidase from influenza virus A/Shandong/9/1993(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Lei, R, Hernandez Garcia, A. | | Deposit date: | 2022-02-28 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prevalence and mechanisms of evolutionary contingency in human influenza H3N2 neuraminidase.
Nat Commun, 13, 2022
|
|
3CLL
 
 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E mutant | | Descriptor: | Aquaporin | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Trnroth-Horsefield, S. | | Deposit date: | 2008-03-19 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
8DWB
 
 | |
6V03
 
 | | ELIC-propylammonium complex in POPC-only nanodiscs | | Descriptor: | 3-AMINOPROPANE, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3B8H
 
 | | Structure of the eEF2-ExoA(E546A)-NAD+ complex | | Descriptor: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jorgensen, R, Merrill, A.R. | | Deposit date: | 2007-11-01 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
6M2B
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with S416 | | Descriptor: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2.
Protein Cell, 11, 2020
|
|
7UJ2
 
 | | OspC Type B | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Outer surface protein C | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural Elucidation of a Protective B Cell Epitope on Outer Surface Protein C (OspC) of the Lyme Disease Spirochete, Borreliella burgdorferi.
Mbio, 14, 2023
|
|
7E39
 
 | | SARS-CoV-2 spike in complex with the Ab4 neutralizing antibody (State 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Ab4, Light Chain of Ab4, ... | | Authors: | Liu, C. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Three epitope-distinct human antibodies from RenMab mice neutralize SARS-CoV-2 and cooperatively minimize the escape of mutants.
Cell Discov, 7, 2021
|
|
7UJ6
 
 | | Outer Surface Protein C Type K | | Descriptor: | Outer surface protein C | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Elucidation of a Protective B Cell Epitope on Outer Surface Protein C (OspC) of the Lyme Disease Spirochete, Borreliella burgdorferi.
Mbio, 14, 2023
|
|
7UIJ
 
 | | Structural studies of B5-OspC complex | | Descriptor: | 1,2-ETHANEDIOL, Monoclonal B5 Fab Heavy Chain, Monoclonal B5 Fab Light Chain, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural Elucidation of a Protective B Cell Epitope on Outer Surface Protein C (OspC) of the Lyme Disease Spirochete, Borreliella burgdorferi.
Mbio, 14, 2023
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
