7RHS
 
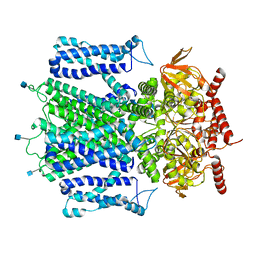 | | Cryo-EM structure of apo-state of human CNGA3/CNGB3 channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cyclic nucleotide-gated cation channel alpha-3, Cyclic nucleotide-gated cation channel beta-3, ... | | Authors: | Zheng, X, Yang, J. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RNM
 
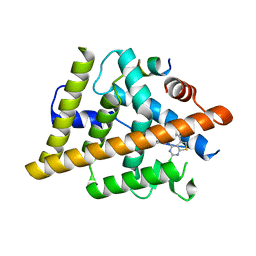 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with 2-(2-Chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)isoindolin-5-ol and GRIP Peptide | | Descriptor: | 2-(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-isoindol-5-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Willson, T.M, Fanning, S.W. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of the Binding and Activation of Estrogen Receptor alpha by Phenolic Thieno[2,3-d]pyrimidines
Helv.Chim.Acta, 106, 2023
|
|
6LUS
 
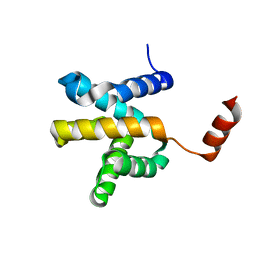 | | Crystal structure of the Mengla Virus VP30 C-terminal domain | | Descriptor: | Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Wen, K.N, Chu, H.G, Wang, C.H, Qin, X.C. | | Deposit date: | 2020-01-30 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Mengla virus VP30 C-terminal domain.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6IMD
 
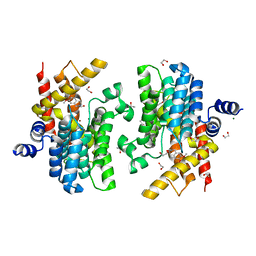 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
8XQ8
 
 | |
8XQ7
 
 | |
8XQ9
 
 | |
8XPQ
 
 | |
8XQA
 
 | |
8XQ4
 
 | |
5BWO
 
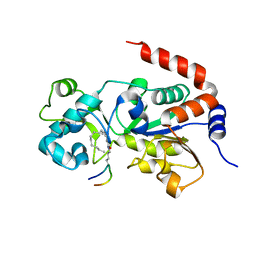 | | Crystal Structure of Human SIRT3 in Complex with a Palmitoyl H3K9 Peptide | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, PALMITIC ACID, ... | | Authors: | Gai, W, Jiang, H, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
2P1W
 
 | |
6IMB
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMI
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-ethoxy-7-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
5D0Q
 
 | | BamACDE complex, outer membrane beta-barrel assembly machinery (BAM) complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, Outer membrane protein assembly factor BamD, ... | | Authors: | Gu, Y, Paterson, N, Zeng, Y, Dong, H, Wang, W, Dong, C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of outer membrane protein insertion by the BAM complex.
Nature, 531, 2016
|
|
5D0O
 
 | | BamABCDE complex, outer membrane beta barrel assembly machinery entire complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Gu, Y, Paterson, N, Zeng, Y, Dong, H, Wang, W, Dong, C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of outer membrane protein insertion by the BAM complex.
Nature, 531, 2016
|
|
3DKK
 
 | | Aged Form of Human Butyrylcholinesterase Inhibited by Tabun | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Carletti, E. | | Deposit date: | 2008-06-25 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
3L5T
 
 | |
5VNA
 
 | | Crystal structure of human YEATS domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
3L5R
 
 | |
7CF8
 
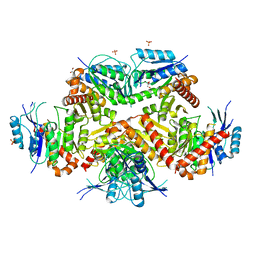 | | PfkB(Mycobacterium marinum) | | Descriptor: | Fructokinase, PfkB, GLYCEROL, ... | | Authors: | Li, J, Gao, B, Ji, R. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
