2YEV
 
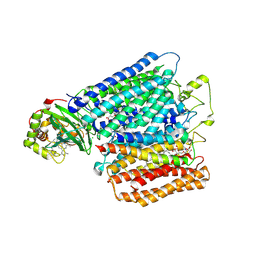 | | Structure of caa3-type cytochrome oxidase | | Descriptor: | (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE, (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, (2R)-3-HYDROXYPROPANE-1,2-DIYL DIHEXADECANOATE, ... | | Authors: | Lyons, J.A, Aragao, D, Soulimane, T, Caffrey, M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Insights Into Electron Transfer in Caa3-Type Cytochrome Oxidases.
Nature, 487, 2012
|
|
2Y9H
 
 | |
1T3L
 
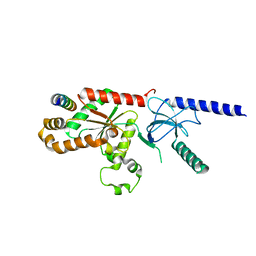 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core in Complex with Alpha1 Interaction Domain | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, Voltage-dependent L-type calcium channel alpha-1S subunit | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Voltage-Dependent Calcium Channel Beta Subunit Functional Core and Its Complex with the Alpha1 Interaction Domain
NEURON, 42, 2004
|
|
1SSP
 
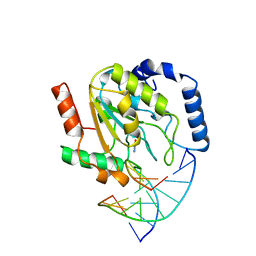 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
3C7G
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with xylotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7E
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3CAY
 
 | | Crystal structure of Lipopeptide Detergent (LPD-12) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LPD-12 | | Authors: | Ho, D.N, Pomroy, N.C, Cuesta-Seijo, J.A, Prive, G.G. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a self-assembling lipopeptide detergent at 1.20 A.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1T3S
 
 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, MERCURY (II) ION | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain.
Neuron, 42, 2004
|
|
3CB3
 
 | | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate | | Descriptor: | L-GLUCARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of L-Talarate dehydratase from Polaromonas sp. JS666 complexed with Mg and L-glucarate.
To be Published
|
|
3C7H
 
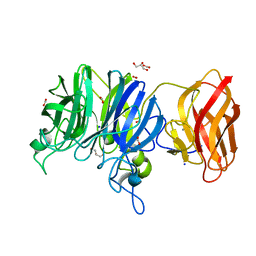 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3CBA
 
 | |
372D
 
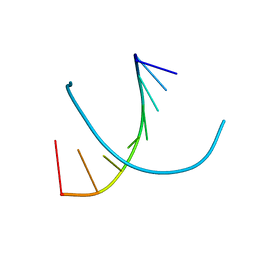 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
397D
 
 | |
362D
 
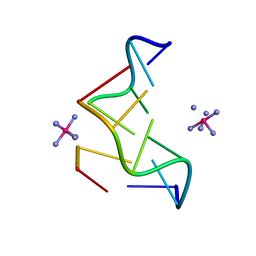 | | THE STRUCTURE OF D(TGCGCA)2 AND A COMPARISON TO OTHER Z-DNA HEXAMERS | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*TP*GP*CP*GP*CP*A)-3') | | Authors: | Harper, N.A, Brannigan, J.A, Buck, M, Lewis, R.J, Moore, M.H, Schneider, B. | | Deposit date: | 1997-08-20 | | Release date: | 1997-11-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of d(TGCGCA)2 and a comparison to other DNA hexamers.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
390D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3') | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
3CF4
 
 | | Structure of the CODH component of the M. barkeri ACDS complex | | Descriptor: | ACETIC ACID, Acetyl-CoA decarboxylase/synthase alpha subunit, Acetyl-CoA decarboxylase/synthase epsilon subunit, ... | | Authors: | Gong, W, Hao, B, Wei, Z, Ferguson Jr, D.J, Tallant, T, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2008-03-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the alpha2 epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarboxylase/synthase complex
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CT2
 
 | | Crystal structure of muconate cycloisomerase from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes.
Biochemistry, 48, 2009
|
|
3CYU
 
 | | Human Carbonic Anhydrase II complexed with Cryptophane biosensor and xenon | | Descriptor: | Carbonic anhydrase 2, MoMo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-y l)propanoic acid]-cryptophane-A, PoPo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-yl)propanoic acid]-cryptophane-A, ... | | Authors: | Aaron, J.A, Jude, K.M, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2008-04-26 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a 129Xe-cryptophane biosensor complexed with human carbonic anhydrase II.
J.Am.Chem.Soc., 130, 2008
|
|
3D47
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate | | Descriptor: | D-MALATE, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate.
To be Published
|
|
3D5J
 
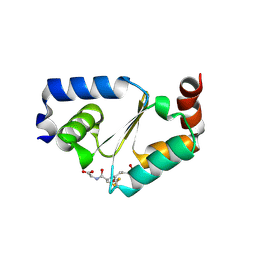 | | Structure of yeast Grx2-C30S mutant with glutathionyl mixed disulfide | | Descriptor: | GLUTATHIONE, Glutaredoxin-2, mitochondrial | | Authors: | Discola, K.F, de Oliveira, M.A, Barcena, J.A, Porras, P, Padilla, C.A, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae.
J.Mol.Biol., 385, 2009
|
|
3CQQ
 
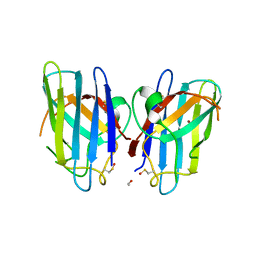 | | Human SOD1 G85R Variant, Structure II | | Descriptor: | ACETYL GROUP, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Cao, X, Antonyuk, S, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
3B5T
 
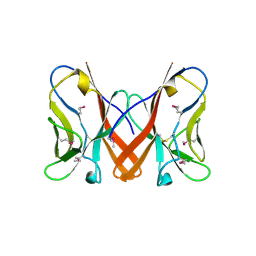 | | Crystal Structure of Novel Immune-Type Receptor 10 Se-Met Extracellular Fragment Mutant N30D | | Descriptor: | Novel immune-type receptor 10 | | Authors: | Ostrov, D.A, Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Magis, A.T, Bailey, K.M, Litman, G.W. | | Deposit date: | 2007-10-26 | | Release date: | 2008-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition.
Immunity, 29, 2008
|
|
3BHT
 
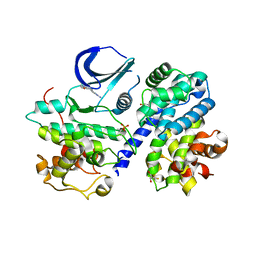 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor meriolin 3 | | Descriptor: | 4-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
3CXO
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and 3-deoxy-L-rhamnonate | | Descriptor: | (2R,4S)-2,4,7-trihydroxyheptanoic acid, 3,6-dideoxy-L-arabino-hexonic acid, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Hubbard, B.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-24 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
3D46
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate | | Descriptor: | L(+)-TARTARIC ACID, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate.
To be Published
|
|
