2JEO
 
 | | Crystal structure of human uridine-cytidine kinase 1 | | Descriptor: | URIDINE-CYTIDINE KINASE 1 | | Authors: | Kosinska, U, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E.P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Uridine-Cytidine Kinase 1
To be Published
|
|
2JBH
 
 | | Human phosphoribosyl transferase domain containing 1 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Welin, M, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Eklund, H, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of the Human Phosphoribosyltransferase Domain Containing Protein 1.
FEBS J., 277, 2010
|
|
2JGY
 
 | | The crystal structure of human orotidine-5'-decarboxylase domain of human uridine monophosphate synthetase (UMPS) | | Descriptor: | UMP SYNTHASE | | Authors: | Moche, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Uppenberg, J, Upsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Human Orotidine-5'-Decarboxylase Domain of Human Uridine Monophosphate Synthetase (Umps)
To be Published
|
|
2JI4
 
 | | Human phosphoribosylpyrophosphate synthetase - associated protein 41 (PAP41) | | Descriptor: | CHLORIDE ION, PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE-ASSOCIATED PROTEIN 2 | | Authors: | Moche, M, Lehtio, L, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Karlberg, T, Kotenyova, T, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human Phosphoribosylpyrophosphate Synthetase -Associated Protein 41 (Pap41)
To be Published
|
|
2L49
 
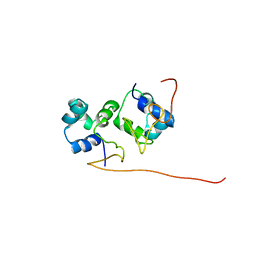 | | The solution structure of the P2 C,the immunity repressor of the P2 bacteriophage | | Descriptor: | C protein | | Authors: | Massad, T, Papadopolos, E, Stenmark, P, Damberg, P. | | Deposit date: | 2010-10-01 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The C repressor of the P2 bacteriophage.
J.Biomol.Nmr, 64, 2016
|
|
5MZF
 
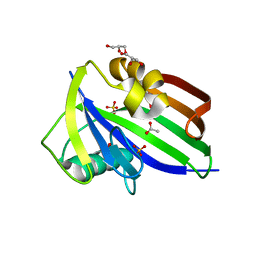 | | Crystal structure of dog MTH1 protein | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Narwal, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
5N0C
 
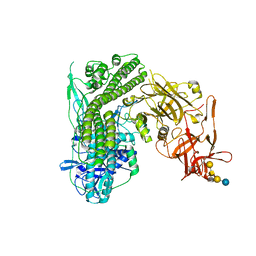 | | Crystal structure of the tetanus neurotoxin in complex with GM1a | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tetanus toxin, ZINC ION, ... | | Authors: | Masuyer, G, Conrad, J, Stenmark, P. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the tetanus toxin reveals pH-mediated domain dynamics.
EMBO Rep., 18, 2017
|
|
5MZG
 
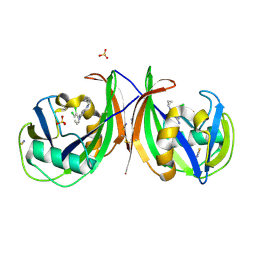 | | Crystal structure of mouse MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION, ... | | Authors: | Narwal, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
5NGR
 
 | | Crystal structure of human MTH1 in complex with fragment inhibitor 8-(methylsulfanyl)-7H-purin-6-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-methylsulfanyl-7~{H}-purin-6-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
5NGS
 
 | | Crystal structure of human MTH1 in complex with inhibitor 6-[(2-phenylethyl)sulfanyl]-7H-purin-2-amine | | Descriptor: | 6-(2-phenylethylsulfanyl)-7~{H}-purin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
5N0B
 
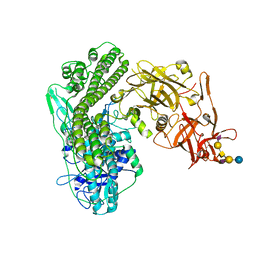 | | Crystal structure of the tetanus neurotoxin in complex with GD1a | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Tetanus toxin, ... | | Authors: | Masuyer, G, Conrad, J, Stenmark, P. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the tetanus toxin reveals pH-mediated domain dynamics.
EMBO Rep., 18, 2017
|
|
5MZE
 
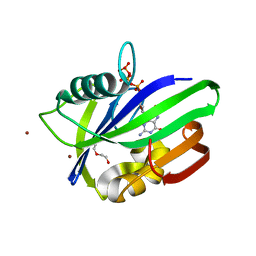 | | Crystal structure of mouse MTH1 with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, COPPER (II) ION, ... | | Authors: | Narwal, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
5NGT
 
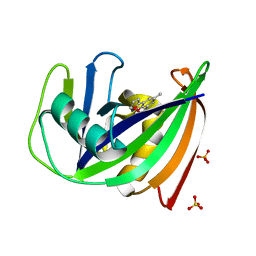 | | Crystal structure of human MTH1 in complex with inhibitor 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 7-(furan-2-yl)-5-methyl-1,3-benzoxazol-2-amine, SULFATE ION | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
5OTN
 
 | | Crystal structure of zebrafish MTH1 in complex with O6-methyl-dGMP | | Descriptor: | 1,2-ETHANEDIOL, 6-O-METHYL GUANOSINE-5'-MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Gustafsson, R, Henriksson, L, Jemth, A.-S, Brautigam, L, Carreras Puigvert, J, Homan, E, Warpman Berglund, U, Helleday, T, Stenmark, P. | | Deposit date: | 2017-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | MutT homologue 1 (MTH1) catalyzes the hydrolysis of mutagenic O6-methyl-dGTP.
Nucleic Acids Res., 46, 2018
|
|
5OTM
 
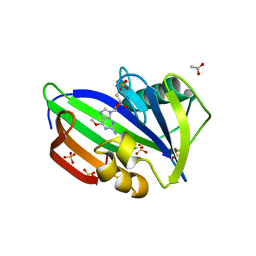 | | Crystal structure of human MTH1 in complex with O6-methyl-dGMP | | Descriptor: | 6-O-METHYL GUANOSINE-5'-MONOPHOSPHATE, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Henriksson, L, Jemth, A.-S, Brautigam, L, Carreras Puigvert, J, Homan, E, Warpman Berglund, U, Helleday, T, Stenmark, P. | | Deposit date: | 2017-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MutT homologue 1 (MTH1) catalyzes the hydrolysis of mutagenic O6-methyl-dGTP.
Nucleic Acids Res., 46, 2018
|
|
6ZJC
 
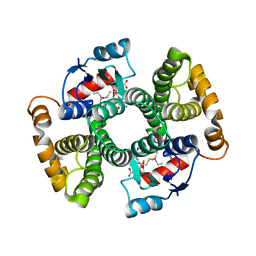 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione and triethyltin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6ZJ9
 
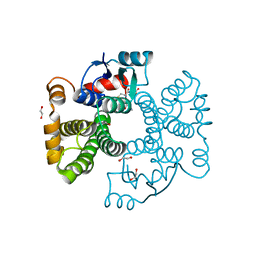 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6ZVM
 
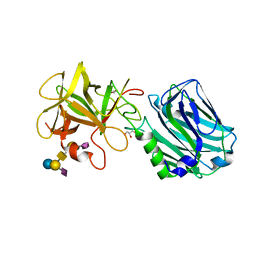 | | Botulinum neurotoxin B2 binding domain in complex with GD1a | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin | | Authors: | Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of Botulinum Neurotoxin Subtype B2 Binding to Its Receptors.
Toxins, 12, 2020
|
|
6ZVN
 
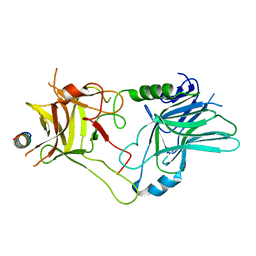 | |
7B67
 
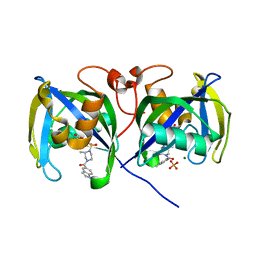 | | Structure of NUDT15 V18_V19insGV Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B65
 
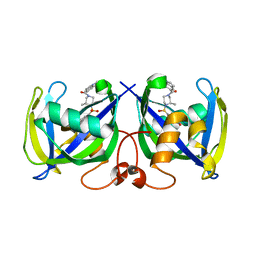 | | Structure of NUDT15 R139C Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B63
 
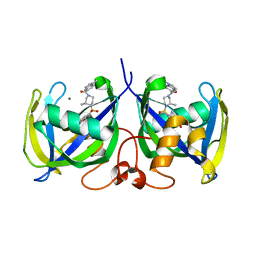 | | Structure of NUDT15 in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B66
 
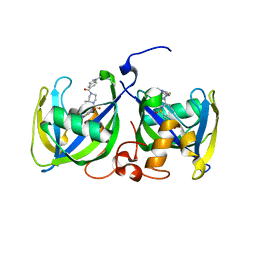 | | Structure of NUDT15 R139H Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B64
 
 | | Structure of NUDT15 V18I Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B7V
 
 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
