1H2U
 
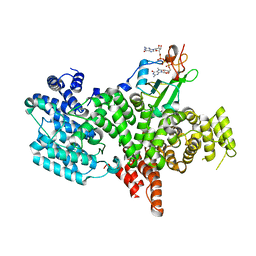 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1H2T
 
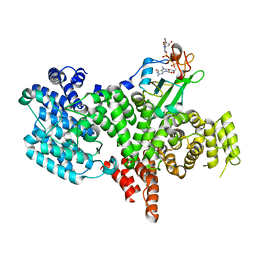 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
6GGS
 
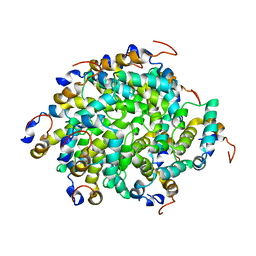 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
6GFJ
 
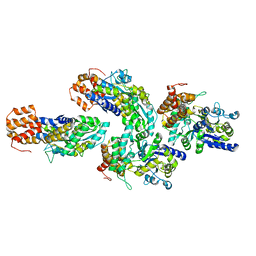 | | Structure of RIP2 CARD domain fused to crystallisable MBP tag | | Descriptor: | Sugar ABC transporter substrate-binding protein,Receptor-interacting serine/threonine-protein kinase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
5FML
 
 | | Crystal structure of the endonuclease from the PA subunit of influenza B virus bound to the PB2 subunit NLS peptide | | Descriptor: | GLYCEROL, MAGNESIUM ION, PA SUBUNIT OF INFLUENZA B POLYMERASE, ... | | Authors: | Guilligay, D, Gaudon, S, Cusack, S. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
1JID
 
 | | Human SRP19 in complex with helix 6 of Human SRP RNA | | Descriptor: | HELIX 6 OF HUMAN SRP RNA, MAGNESIUM ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN | | Authors: | Wild, K, Sinning, I, Cusack, S. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an early protein-RNA assembly complex of the signal recognition particle.
Science, 294, 2001
|
|
7PQK
 
 | |
1LZW
 
 | | Structural basis of ClpS-mediated switch in ClpA substrate recognition | | Descriptor: | ATP-dependent clp protease ATP-binding subunit ClpA, PLATINUM (II) ION, Protein yljA | | Authors: | Zeth, K, Ravelli, R.B, Paal, K, Cusack, S, Bukau, B, Dougan, D.A. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA
Nat.Struct.Biol., 9, 2002
|
|
5FMM
 
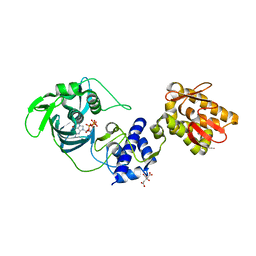 | | crystal structure of the mid, cap-binding, mid-link and 627 domains from avian influenza a virus polymerase PB2 subunit bound to M7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CITRATE ANION, INFLUENZA A PB2 SUBUNIT | | Authors: | Thierry, E, Pflug, A, Hart, D, Cusack, S. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
5FMQ
 
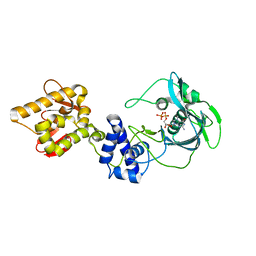 | | Crystal structure of the mid, cap-binding, mid-link and 627 domains from avian influenza A virus polymerase PB2 subunit bound to M7GTP H32 crystal form | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, INFLUENZA A PB2 SUBUNIT | | Authors: | Thierry, E, Pflug, A, Hart, D, Cusack, S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
5AH5
 
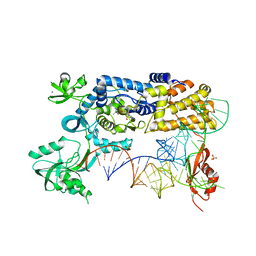 | | Crystal structure of the ternary complex of Agrobacterium radiobacter K84 agnB2 LeuRS-tRNA-LeuAMS | | Descriptor: | 5'-O-(L-leucylsulfamoyl)adenosine, LEUCINE--TRNA LIGASE, MANGANESE (II) ION, ... | | Authors: | Palencia, A, Chopra, S, Virus, C, Schulwitz, S, Temple, B.R, Cusack, S, Reader, J.S. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Antibiotic Self-Immunity tRNA Synthetase in Plant Tumour Biocontrol Agent.
Nat.Commun., 7, 2016
|
|
1MG9
 
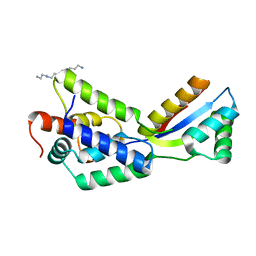 | | The structural basis of ClpS-mediated switch in ClpA substrate recognition | | Descriptor: | ATP dependent clp protease ATP-binding subunit clpA, SPERMINE (FULLY PROTONATED FORM), protein yljA | | Authors: | Zeth, K, Ravelli, R.B, Paal, K, Cusack, S, Bukau, B, Dougan, D.A. | | Deposit date: | 2002-08-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA
Nat.Struct.Biol., 9, 2002
|
|
5MV0
 
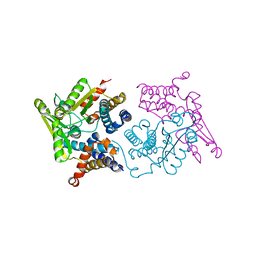 | | Structure of an N-terminal domain of a reptarenavirus L protein | | Descriptor: | L protein, PHOSPHATE ION | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
5MUZ
 
 | | Structure of a C-terminal domain of a reptarenavirus L protein | | Descriptor: | L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
1QHV
 
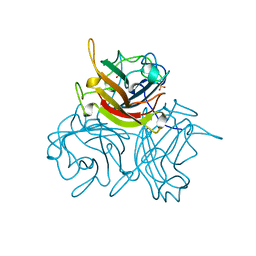 | | HUMAN ADENOVIRUS SEROTYPE 2 FIBRE HEAD | | Descriptor: | PROTEIN (ADENOVIRUS FIBRE), SULFATE ION | | Authors: | Van Raaij, M.J, Louis, N, Chroboczek, J, Cusack, S. | | Deposit date: | 1999-05-28 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the human adenovirus serotype 2 fiber head domain at 1.5 A resolution.
Virology, 262, 1999
|
|
1QIU
 
 | |
5MUY
 
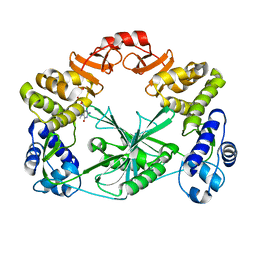 | | Structure of a C-terminal domain of a reptarenavirus L protein with m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
2XGT
 
 | | Asparaginyl-tRNA synthetase from Brugia malayi complexed with the sulphamoyl analogue of asparaginyl-adenylate | | Descriptor: | 5'-O-[N-(L-ASPARAGINYL)SULFAMOYL]ADENOSINE, ASPARAGINYL-TRNA SYNTHETASE, CYTOPLASMIC, ... | | Authors: | Crepin, T, Haertlein, M, Kron, M, Cusack, S. | | Deposit date: | 2010-06-07 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Hybrid Structural Model of the Complete Brugia Malayi Cytoplasmic Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 405, 2011
|
|
2XTI
 
 | | Asparaginyl-tRNA synthetase from Brugia malayi complexed with ATP:Mg and L-Asp-beta-NOH adenylate:PPi:Mg | | Descriptor: | 5'-O-[(R)-{[(2S)-2-amino-4-(hydroxyamino)-4-oxobutanoyl]oxy}(hydroxy)phosphoryl]adenosine, ADENOSINE-5'-TRIPHOSPHATE, ASPARAGINYL-TRNA SYNTHETASE, ... | | Authors: | Crepin, T, Haertlein, M, Kron, M, Cusack, S. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Hybrid Structural Model of the Complete Brugia Malayi Cytoplasmic Asparaginyl-tRNA Synthetase.
J.Mol.Biol., 405, 2011
|
|
8PM0
 
 | |
1E8O
 
 | | Core of the Alu domain of the mammalian SRP | | Descriptor: | 7SL RNA, SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Weichenrieder, O, Wild, K, Strub, K, Cusack, S. | | Deposit date: | 2000-09-28 | | Release date: | 2000-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Assembly of the Alu Domain of the Mammalian Signal Recognition Particle
Nature, 408, 2000
|
|
1EAJ
 
 | |
8R3K
 
 | |
5AGJ
 
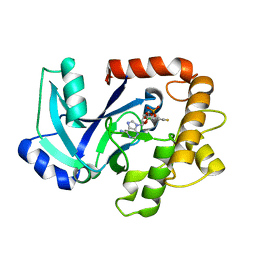 | | Crystal structure of the LeuRS editing domain of Candida albicans in complex with the adduct AN2690-AMP | | Descriptor: | POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
5AGI
 
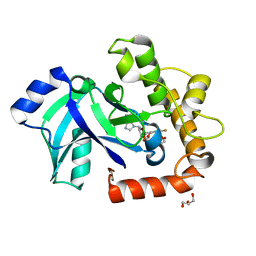 | | Crystal structure of the LeuRS editing domain of Candida albicans Mutant K510A in complex with the adduct formed by AN2690-AMP | | Descriptor: | GLYCEROL, POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
