3E53
 
 | |
6JT9
 
 | | Crystal Structure of D464A mutant of FGAM Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6JTA
 
 | | Crystal Structure of D464A L465A mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6JT7
 
 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6JT8
 
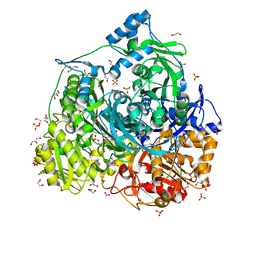 | | Crystal structure of 450-451_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
5FIQ
 
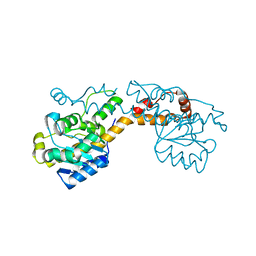 | | Exonuclease domain-containing 1 (Exd1) in the native conformation | | Descriptor: | EXD1 | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
5H58
 
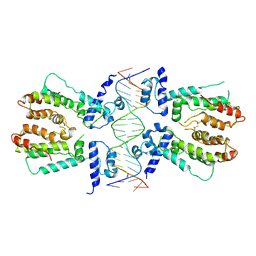 | | Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence | | Descriptor: | CprB, DNA (5'-D(*AP*GP*GP*C*AP*GP*GP*CP*GP*GP*CP*AP*CP*GP*GP*TP*CP*TP*GP*TP*TP*GP*AP*GP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*A*CP*TP*CP*AP*AP*CP*AP*GP*AP*CP*CP*GP*TP*GP*CP*CP*GP*CP*CP*TP*GP*CP*CP*T)-3') | | Authors: | Bhukya, H, Jana, A.K, Sengupta, N, Anand, R. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (3.991 Å) | | Cite: | Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence
J. Struct. Biol., 198, 2017
|
|
5H4E
 
 | |
3PD5
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with an analog of threonyl-adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, GLYCEROL, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kamarthapu, V, Kruparani, S.P, Sankaranarayanan, R. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mechanistic insights into cognate substrate discrimination during proofreading in translation
Proc.Natl.Acad.Sci.USA, 2010
|
|
5HT9
 
 | |
5FIS
 
 | | Exonuclease domain-containing 1 (Exd1) in the Gd bound conformation | | Descriptor: | EXD1, GADOLINIUM ATOM | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-02 | | Release date: | 2015-12-23 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
3PD4
 
 | |
3PD3
 
 | |
3PD2
 
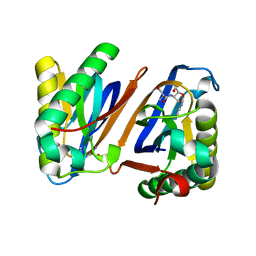 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kamarthapu, V, Kruparani, S.P, Sankaranarayanan, R. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanistic insights into cognate substrate discrimination during proofreading in translation
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5E7V
 
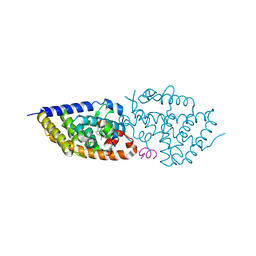 | | Potent Vitamin D Receptor Agonist | | Descriptor: | 1-ALPHA-HYDROXY-27-NOR-25-O-CARBONYL-VITAMIN D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Otero, R, Seoane, S, Sigueiro, R, Belorusova, A.Y, Maestro, M.A, Perez-Fernandez, R, Rochel, N, Mourino, A. | | Deposit date: | 2015-10-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carborane-based design of a potent vitamin D receptor agonist.
Chem Sci, 7, 2016
|
|
5A3F
 
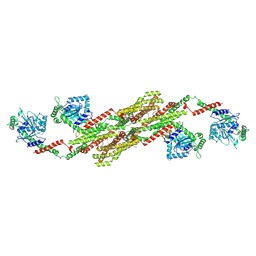 | | Crystal structure of the dynamin tetramer | | Descriptor: | DYNAMIN 3 | | Authors: | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
3SNZ
 
 | |
3BT4
 
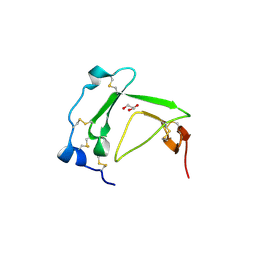 | | Crystal Structure Analysis of AmFPI-1, fungal protease inhibitor from Antheraea mylitta | | Descriptor: | Fungal protease inhibitor-1, GLYCEROL | | Authors: | Roy, S, Aravind, P, Madhurantakam, C, Ghosh, A.K, Sankarananarayanan, R, Das, A.K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a fungal protease inhibitor from Antheraea mylitta
J.Struct.Biol., 166, 2009
|
|
3SNY
 
 | |
3SO1
 
 | |
4JFM
 
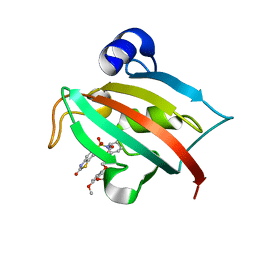 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate | | Descriptor: | 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
3SO0
 
 | |
4JFI
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione | | Descriptor: | 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFJ
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFK
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
